Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
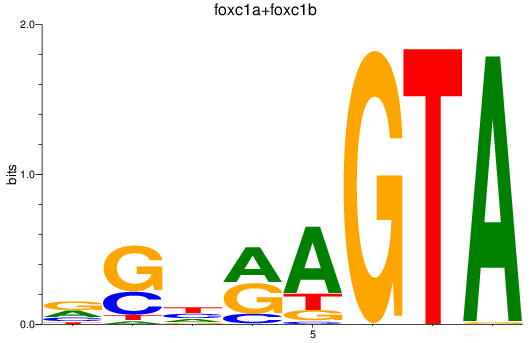
Results for foxc1a+foxc1b
Z-value: 0.82
Transcription factors associated with foxc1a+foxc1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxc1b
|
ENSDARG00000055398 | forkhead box C1b |
|
foxc1a
|
ENSDARG00000091481 | forkhead box C1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxc1a | dr11_v1_chr2_-_689047_689047 | -0.47 | 4.4e-02 | Click! |
| foxc1b | dr11_v1_chr20_+_26702377_26702377 | -0.16 | 5.1e-01 | Click! |
Activity profile of foxc1a+foxc1b motif
Sorted Z-values of foxc1a+foxc1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_46310204 | 3.54 |
ENSDART00000188713
ENSDART00000184930 |
crybb1l1
|
crystallin, beta B1, like 1 |
| chr22_+_20720808 | 2.87 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr2_-_22152797 | 2.24 |
ENSDART00000145188
|
cyp7a1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr21_-_43606502 | 2.23 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr1_-_22338521 | 2.21 |
ENSDART00000176849
|
si:ch73-380n15.2
|
si:ch73-380n15.2 |
| chr12_-_22524388 | 2.13 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr7_+_23907692 | 2.08 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr15_-_12319065 | 2.07 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr10_+_2587234 | 1.96 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr16_-_13818061 | 1.95 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr9_-_48736388 | 1.94 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr9_-_7421135 | 1.91 |
ENSDART00000144600
|
bin1a
|
bridging integrator 1a |
| chr21_-_4032650 | 1.78 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr25_+_20216159 | 1.73 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr8_+_53464216 | 1.71 |
ENSDART00000169514
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr15_+_47903864 | 1.61 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr4_-_4387012 | 1.58 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr1_-_54947592 | 1.56 |
ENSDART00000129710
|
crtac1a
|
cartilage acidic protein 1a |
| chr21_-_22114625 | 1.53 |
ENSDART00000177426
ENSDART00000135410 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr5_+_72087619 | 1.47 |
ENSDART00000062885
|
oxt
|
oxytocin |
| chr13_+_24552254 | 1.43 |
ENSDART00000147907
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr1_+_7540978 | 1.43 |
ENSDART00000147770
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr17_-_12336987 | 1.36 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr22_-_10486477 | 1.33 |
ENSDART00000184366
|
aspn
|
asporin (LRR class 1) |
| chr22_-_10487490 | 1.31 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr23_+_6272638 | 1.29 |
ENSDART00000190366
|
syt2a
|
synaptotagmin IIa |
| chr22_+_5103349 | 1.27 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr19_-_19339285 | 1.25 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr11_+_30729745 | 1.23 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr1_-_45177373 | 1.22 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr8_+_19514294 | 1.22 |
ENSDART00000170622
|
si:ch73-281k2.5
|
si:ch73-281k2.5 |
| chr23_-_24856025 | 1.21 |
ENSDART00000142171
|
syt6a
|
synaptotagmin VIa |
| chr5_+_30680804 | 1.16 |
ENSDART00000078049
|
pdzd3b
|
PDZ domain containing 3b |
| chr2_+_20430366 | 1.13 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr19_-_5103313 | 1.10 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr1_-_8101495 | 1.06 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr11_+_11200550 | 1.05 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr4_-_53370 | 1.03 |
ENSDART00000180254
ENSDART00000186529 |
CU856344.2
|
|
| chr16_-_42004544 | 1.00 |
ENSDART00000034544
|
caspa
|
caspase a |
| chr1_-_14332283 | 0.99 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr24_-_21588914 | 0.98 |
ENSDART00000152054
|
gpr12
|
G protein-coupled receptor 12 |
| chr23_+_18944388 | 0.97 |
ENSDART00000104487
|
cox4i2
|
cytochrome c oxidase subunit IV isoform 2 |
| chr18_-_41219790 | 0.95 |
ENSDART00000162387
ENSDART00000193753 |
zbtb38
|
zinc finger and BTB domain containing 38 |
| chr3_-_40976463 | 0.93 |
ENSDART00000128450
ENSDART00000018676 |
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr22_+_31930650 | 0.93 |
ENSDART00000092090
|
dock3
|
dedicator of cytokinesis 3 |
| chr9_-_7257877 | 0.92 |
ENSDART00000153514
|
si:ch211-121j5.4
|
si:ch211-121j5.4 |
| chr12_-_26064480 | 0.92 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr18_-_12858016 | 0.91 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr15_-_568645 | 0.91 |
ENSDART00000156744
|
cbln18
|
cerebellin 18 |
| chr25_-_31739309 | 0.91 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr13_-_301309 | 0.85 |
ENSDART00000131747
|
chs1
|
chitin synthase 1 |
| chr12_+_10116912 | 0.85 |
ENSDART00000189630
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr24_-_32408404 | 0.83 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr16_+_13818500 | 0.81 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr3_-_18030938 | 0.81 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr2_-_50372467 | 0.78 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr7_-_38790341 | 0.77 |
ENSDART00000159884
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr8_-_46525092 | 0.76 |
ENSDART00000030482
|
sult1st2
|
sulfotransferase family 1, cytosolic sulfotransferase 2 |
| chr8_-_15398760 | 0.75 |
ENSDART00000162011
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr5_+_25952340 | 0.73 |
ENSDART00000147188
|
trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr12_-_24060358 | 0.72 |
ENSDART00000050831
|
chac2
|
ChaC, cation transport regulator homolog 2 (E. coli) |
| chr15_-_37921998 | 0.69 |
ENSDART00000193597
ENSDART00000181443 ENSDART00000168790 |
si:dkey-238d18.5
|
si:dkey-238d18.5 |
| chr13_+_36680564 | 0.68 |
ENSDART00000136030
ENSDART00000006923 |
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex subunit s |
| chr17_+_53250802 | 0.68 |
ENSDART00000143819
|
VASH1
|
vasohibin 1 |
| chr21_+_11105007 | 0.68 |
ENSDART00000128859
ENSDART00000193105 |
prlra
|
prolactin receptor a |
| chr20_-_3166168 | 0.67 |
ENSDART00000134137
|
si:ch73-212j7.3
|
si:ch73-212j7.3 |
| chr16_-_12723324 | 0.67 |
ENSDART00000131915
|
sbk3
|
SH3 domain binding kinase family, member 3 |
| chr12_-_26064105 | 0.67 |
ENSDART00000168825
|
ldb3b
|
LIM domain binding 3b |
| chr1_-_7917062 | 0.67 |
ENSDART00000177068
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr7_-_28463106 | 0.67 |
ENSDART00000137799
|
trim66
|
tripartite motif containing 66 |
| chr14_+_35013656 | 0.65 |
ENSDART00000164974
|
ebf3a
|
early B cell factor 3a |
| chr20_-_27311675 | 0.65 |
ENSDART00000026088
ENSDART00000148361 |
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr9_+_12890161 | 0.63 |
ENSDART00000146477
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr9_-_23994225 | 0.62 |
ENSDART00000140346
|
col6a3
|
collagen, type VI, alpha 3 |
| chr21_-_26250744 | 0.61 |
ENSDART00000044735
|
gria1b
|
glutamate receptor, ionotropic, AMPA 1b |
| chr4_+_57093908 | 0.60 |
ENSDART00000170198
|
si:ch211-238e22.5
|
si:ch211-238e22.5 |
| chr18_+_808911 | 0.60 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr8_-_44015210 | 0.59 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr1_+_6817292 | 0.59 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr5_-_28029558 | 0.59 |
ENSDART00000078649
|
abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr15_+_15456910 | 0.59 |
ENSDART00000155708
|
zgc:174895
|
zgc:174895 |
| chr8_-_18200003 | 0.59 |
ENSDART00000080014
|
rps8b
|
ribosomal protein S8b |
| chr3_-_40976288 | 0.57 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr3_-_43770876 | 0.57 |
ENSDART00000160162
|
zgc:92162
|
zgc:92162 |
| chr24_-_25428176 | 0.57 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr9_-_3866707 | 0.56 |
ENSDART00000010378
|
myo3b
|
myosin IIIB |
| chr13_-_47800342 | 0.56 |
ENSDART00000121817
|
fbln7
|
fibulin 7 |
| chr5_+_23597144 | 0.56 |
ENSDART00000143929
ENSDART00000142420 ENSDART00000032909 |
kat5b
|
K(lysine) acetyltransferase 5b |
| chr17_-_465285 | 0.56 |
ENSDART00000168718
|
chrm5a
|
cholinergic receptor, muscarinic 5a |
| chr3_-_16227683 | 0.56 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr23_-_44819100 | 0.55 |
ENSDART00000076373
|
st8sia7.1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 7.1 |
| chr20_-_46817223 | 0.55 |
ENSDART00000100336
|
esrrgb
|
estrogen-related receptor gamma b |
| chr7_-_22132265 | 0.52 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr19_+_164013 | 0.51 |
ENSDART00000167379
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr1_-_45341760 | 0.50 |
ENSDART00000149183
ENSDART00000148289 ENSDART00000110390 |
zgc:101679
|
zgc:101679 |
| chr7_-_31321027 | 0.49 |
ENSDART00000186878
|
CR356242.1
|
|
| chr21_+_6556635 | 0.49 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr2_+_6181383 | 0.47 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr5_-_67629263 | 0.46 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr11_-_29946927 | 0.46 |
ENSDART00000165182
|
zgc:113276
|
zgc:113276 |
| chr23_-_42752387 | 0.45 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr1_-_25679339 | 0.45 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr1_-_46326156 | 0.44 |
ENSDART00000040284
|
atp11a
|
ATPase phospholipid transporting 11A |
| chr19_-_10730488 | 0.43 |
ENSDART00000126033
|
slc9a3.1
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 1 |
| chr5_-_61942482 | 0.42 |
ENSDART00000097338
|
napaa
|
N-ethylmaleimide-sensitive factor attachment protein, alpha a |
| chr7_+_49695904 | 0.41 |
ENSDART00000183550
ENSDART00000126991 |
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr1_-_37383741 | 0.41 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr7_-_24373662 | 0.41 |
ENSDART00000173865
|
PTGR1 (1 of many)
|
si:dkey-11k2.7 |
| chr6_+_18359306 | 0.40 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr5_-_23705828 | 0.40 |
ENSDART00000189419
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr10_+_15107886 | 0.40 |
ENSDART00000188047
ENSDART00000164095 |
scpp8
|
secretory calcium-binding phosphoprotein 8 |
| chr22_-_26175237 | 0.40 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr22_-_20386982 | 0.40 |
ENSDART00000089015
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr20_+_38276690 | 0.39 |
ENSDART00000061437
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr8_-_29706882 | 0.39 |
ENSDART00000045909
|
prf1.5
|
perforin 1.5 |
| chr7_+_9981757 | 0.36 |
ENSDART00000113429
ENSDART00000173233 |
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr4_+_12333168 | 0.34 |
ENSDART00000172455
ENSDART00000146166 |
pimr214
pimr171
|
Pim proto-oncogene, serine/threonine kinase, related 214 Pim proto-oncogene, serine/threonine kinase, related 171 |
| chr3_-_16227490 | 0.34 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr23_+_14269311 | 0.34 |
ENSDART00000170414
|
CR354537.1
|
|
| chr9_+_51180508 | 0.33 |
ENSDART00000138990
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr5_-_18897482 | 0.33 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr8_-_36370552 | 0.33 |
ENSDART00000097932
ENSDART00000148323 |
si:busm1-104n07.3
|
si:busm1-104n07.3 |
| chr22_-_23000815 | 0.32 |
ENSDART00000137111
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr6_+_42587637 | 0.31 |
ENSDART00000179964
|
camkva
|
CaM kinase-like vesicle-associated a |
| chr12_-_35095414 | 0.31 |
ENSDART00000153229
|
si:dkey-21e13.3
|
si:dkey-21e13.3 |
| chr12_+_20641471 | 0.31 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr5_+_31860043 | 0.31 |
ENSDART00000036235
ENSDART00000140541 |
iscub
|
iron-sulfur cluster assembly enzyme b |
| chr13_+_21601149 | 0.31 |
ENSDART00000179369
|
sh2d4ba
|
SH2 domain containing 4Ba |
| chr22_+_29135846 | 0.30 |
ENSDART00000143811
|
baiap2l2b
|
BAI1-associated protein 2-like 2b |
| chr22_-_7778895 | 0.29 |
ENSDART00000125489
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr13_+_28417297 | 0.28 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr16_-_9383629 | 0.28 |
ENSDART00000084264
ENSDART00000166958 |
adcy2a
|
adenylate cyclase 2a |
| chr17_-_33415740 | 0.28 |
ENSDART00000135218
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr6_+_52853384 | 0.27 |
ENSDART00000174155
ENSDART00000164391 |
BX957318.1
|
|
| chr22_+_24559947 | 0.27 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr3_+_29641181 | 0.27 |
ENSDART00000151517
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr25_+_19238175 | 0.26 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr5_-_4219741 | 0.26 |
ENSDART00000100093
|
si:ch211-283g2.2
|
si:ch211-283g2.2 |
| chr22_-_8860869 | 0.25 |
ENSDART00000166188
|
CABZ01046427.2
|
|
| chr13_+_8696825 | 0.25 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr5_+_37890521 | 0.25 |
ENSDART00000140207
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr4_-_25064510 | 0.24 |
ENSDART00000025153
|
gata3
|
GATA binding protein 3 |
| chr9_-_7089303 | 0.23 |
ENSDART00000146609
|
coa5
|
cytochrome C oxidase assembly factor 5 |
| chr13_+_21600946 | 0.23 |
ENSDART00000144045
|
sh2d4ba
|
SH2 domain containing 4Ba |
| chr24_-_36270855 | 0.23 |
ENSDART00000154858
|
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr18_-_16792561 | 0.22 |
ENSDART00000145546
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr1_+_18550864 | 0.22 |
ENSDART00000142515
|
si:dkey-192k22.2
|
si:dkey-192k22.2 |
| chr2_+_49644803 | 0.22 |
ENSDART00000160342
|
si:ch211-209f23.7
|
si:ch211-209f23.7 |
| chr21_-_4918735 | 0.22 |
ENSDART00000150936
|
kcnt1
|
potassium channel, subfamily T, member 1 |
| chr7_+_38762043 | 0.22 |
ENSDART00000036461
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr9_+_13985567 | 0.22 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr1_-_17663108 | 0.21 |
ENSDART00000131559
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr5_-_12687357 | 0.21 |
ENSDART00000193206
|
aifm3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr20_+_28960340 | 0.21 |
ENSDART00000147917
|
gpr176
|
G protein-coupled receptor 176 |
| chr1_-_35113974 | 0.21 |
ENSDART00000192811
ENSDART00000167461 |
BX897691.1
|
|
| chr8_+_7737062 | 0.21 |
ENSDART00000166712
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr5_-_54300725 | 0.20 |
ENSDART00000163048
|
slc25a35
|
solute carrier family 25, member 35 |
| chr7_-_16214889 | 0.20 |
ENSDART00000076318
ENSDART00000173946 |
btr06
|
bloodthirsty-related gene family, member 6 |
| chr15_-_36347858 | 0.20 |
ENSDART00000155274
ENSDART00000157936 |
si:dkey-23k10.2
|
si:dkey-23k10.2 |
| chr1_-_37383539 | 0.20 |
ENSDART00000127579
|
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr23_+_2185397 | 0.19 |
ENSDART00000109373
|
c1qtnf7
|
C1q and TNF related 7 |
| chr9_+_18576047 | 0.19 |
ENSDART00000145174
|
lacc1
|
laccase (multicopper oxidoreductase) domain containing 1 |
| chr5_-_50640171 | 0.18 |
ENSDART00000183353
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr1_+_23274710 | 0.18 |
ENSDART00000036448
|
lias
|
lipoic acid synthetase |
| chr16_+_28727383 | 0.18 |
ENSDART00000149753
|
dcst2
|
DC-STAMP domain containing 2 |
| chr1_+_35862550 | 0.17 |
ENSDART00000132118
|
si:ch211-194g2.4
|
si:ch211-194g2.4 |
| chr5_-_57624425 | 0.16 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr6_+_72040 | 0.16 |
ENSDART00000122957
|
crygm4
|
crystallin, gamma M4 |
| chr3_-_21062706 | 0.16 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr15_+_638457 | 0.15 |
ENSDART00000157162
|
znf1013
|
zinc finger protein 1013 |
| chr20_-_19365875 | 0.15 |
ENSDART00000063703
ENSDART00000187707 ENSDART00000161065 |
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr5_-_35201482 | 0.15 |
ENSDART00000166272
|
fcho2
|
FCH domain only 2 |
| chr10_+_23520952 | 0.15 |
ENSDART00000125317
|
zmp:0000000937
|
zmp:0000000937 |
| chr12_-_31794908 | 0.14 |
ENSDART00000105583
ENSDART00000153449 |
nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr19_-_17526735 | 0.14 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr19_-_18667130 | 0.14 |
ENSDART00000164833
|
st3gal1l3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1, like 3 |
| chr8_+_54263946 | 0.13 |
ENSDART00000193119
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr4_+_12358822 | 0.13 |
ENSDART00000172557
ENSDART00000150627 |
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr15_-_6966221 | 0.12 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr9_+_12896340 | 0.12 |
ENSDART00000136417
|
si:ch211-167j6.5
|
si:ch211-167j6.5 |
| chr3_+_25825043 | 0.12 |
ENSDART00000153749
|
pik3r6b
|
phosphoinositide-3-kinase, regulatory subunit 6b |
| chr6_+_9398074 | 0.12 |
ENSDART00000151772
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr4_-_14899113 | 0.12 |
ENSDART00000137829
|
si:dkey-14k9.3
|
si:dkey-14k9.3 |
| chr20_+_51478939 | 0.12 |
ENSDART00000149758
|
tlr5a
|
toll-like receptor 5a |
| chr6_+_18441331 | 0.10 |
ENSDART00000171005
|
si:dkey-31g6.4
|
si:dkey-31g6.4 |
| chr17_+_45472608 | 0.10 |
ENSDART00000103493
ENSDART00000183474 |
zgc:110783
|
zgc:110783 |
| chr21_+_34976600 | 0.09 |
ENSDART00000191672
ENSDART00000139635 |
rbm11
|
RNA binding motif protein 11 |
| chr24_-_10917243 | 0.09 |
ENSDART00000144372
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr22_+_2632961 | 0.09 |
ENSDART00000106278
|
znf1173
|
zinc finger protein 1173 |
| chr6_-_51771634 | 0.09 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr2_-_36914281 | 0.09 |
ENSDART00000008322
|
si:dkey-193b15.5
|
si:dkey-193b15.5 |
| chr25_+_16601839 | 0.09 |
ENSDART00000008986
|
atp6v1e1a
|
ATPase H+ transporting V1 subunit E1a |
| chr5_-_61431809 | 0.08 |
ENSDART00000082952
|
rcc1l
|
RCC1 like |
| chr9_-_370225 | 0.08 |
ENSDART00000163637
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr4_+_12342173 | 0.08 |
ENSDART00000161518
|
pimr214
|
Pim proto-oncogene, serine/threonine kinase, related 214 |
| chr6_-_24384654 | 0.08 |
ENSDART00000164723
|
brdt
|
bromodomain, testis-specific |
| chr2_-_11096489 | 0.08 |
ENSDART00000156753
|
lrrc53
|
leucine rich repeat containing 53 |
| chr16_-_40155424 | 0.07 |
ENSDART00000191763
|
BX470182.2
|
|
| chr6_-_8480815 | 0.07 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr25_-_3087556 | 0.07 |
ENSDART00000193249
|
best1
|
bestrophin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxc1a+foxc1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.3 | 1.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.2 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.2 | 0.5 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.2 | 0.9 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.2 | 0.7 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 3.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 0.8 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 1.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 2.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.2 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.1 | 0.7 | GO:0050951 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.1 | 1.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.8 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 4.6 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 1.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.8 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.6 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 1.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.3 | GO:0032728 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.6 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.5 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 1.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 2.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.6 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.8 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.7 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 1.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0043703 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.0 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 3.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 1.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 1.9 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.8 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 1.7 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.7 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.1 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.0 | 0.1 | GO:0014829 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) vascular smooth muscle contraction(GO:0014829) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.3 | 1.0 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 1.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 2.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 1.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.8 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 1.0 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 5.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.5 | GO:0099503 | secretory vesicle(GO:0099503) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.4 | 1.2 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.3 | 1.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 1.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 2.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 0.6 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.2 | 0.6 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.2 | 0.7 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.7 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 1.7 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 1.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 4.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.8 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 2.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 2.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 1.0 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.3 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.6 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 3.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 3.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.3 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.3 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 2.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |