Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
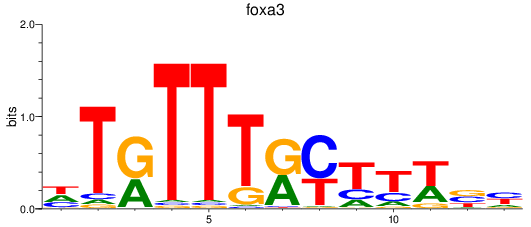
Results for foxa3
Z-value: 1.40
Transcription factors associated with foxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxa3
|
ENSDARG00000012788 | forkhead box A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxa3 | dr11_v1_chr18_-_46354269_46354269 | 0.53 | 2.1e-02 | Click! |
Activity profile of foxa3 motif
Sorted Z-values of foxa3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_36513605 | 3.08 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr5_-_20194876 | 2.85 |
ENSDART00000122587
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr7_-_32020100 | 2.30 |
ENSDART00000185433
|
kif18a
|
kinesin family member 18A |
| chr12_+_27537357 | 2.28 |
ENSDART00000136212
|
etv4
|
ets variant 4 |
| chr3_-_50865079 | 2.20 |
ENSDART00000164295
|
pmp22a
|
peripheral myelin protein 22a |
| chr5_-_54712159 | 2.15 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr18_+_20566817 | 2.05 |
ENSDART00000100716
|
bida
|
BH3 interacting domain death agonist |
| chr17_+_24718272 | 2.03 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr16_-_21785261 | 2.01 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr4_+_14926948 | 1.95 |
ENSDART00000019647
|
psmc2
|
proteasome 26S subunit, ATPase 2 |
| chr24_+_19542323 | 1.93 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr8_+_39795918 | 1.91 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr16_+_38940758 | 1.90 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr2_-_57264262 | 1.85 |
ENSDART00000183815
ENSDART00000149829 ENSDART00000088508 ENSDART00000149508 |
mbd3a
|
methyl-CpG binding domain protein 3a |
| chr4_+_14981854 | 1.84 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr4_-_6373735 | 1.82 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr19_-_43757568 | 1.76 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr16_+_23978978 | 1.75 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr3_+_23743139 | 1.73 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr14_-_16082806 | 1.72 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr20_+_29209615 | 1.68 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr1_+_9153141 | 1.67 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr19_+_22347627 | 1.67 |
ENSDART00000109157
|
fbxl6
|
F-box and leucine-rich repeat protein 6 |
| chr13_+_5978809 | 1.66 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr17_-_42218652 | 1.63 |
ENSDART00000081396
ENSDART00000190007 |
nkx2.2a
|
NK2 homeobox 2a |
| chr4_-_16451375 | 1.63 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr1_+_21309386 | 1.62 |
ENSDART00000054442
ENSDART00000145661 |
med28
|
mediator complex subunit 28 |
| chr11_-_36350421 | 1.60 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr7_-_26844064 | 1.59 |
ENSDART00000162241
|
si:ch211-107p11.3
|
si:ch211-107p11.3 |
| chr11_-_11336986 | 1.58 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr9_-_24030795 | 1.47 |
ENSDART00000144163
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr20_-_9980318 | 1.47 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr7_+_52712807 | 1.45 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr24_+_23716918 | 1.44 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr1_+_11107688 | 1.44 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr19_+_1873059 | 1.42 |
ENSDART00000145246
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr7_-_8417315 | 1.42 |
ENSDART00000173046
|
jac1
|
jacalin 1 |
| chr2_-_30200206 | 1.41 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr1_+_29162478 | 1.39 |
ENSDART00000141441
|
ankrd10a
|
ankyrin repeat domain 10a |
| chr8_+_25902170 | 1.37 |
ENSDART00000193130
|
rhoab
|
ras homolog gene family, member Ab |
| chr8_-_19313510 | 1.37 |
ENSDART00000164780
ENSDART00000137133 |
rgl1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr11_-_36350876 | 1.36 |
ENSDART00000146495
ENSDART00000020655 |
psma5
|
proteasome subunit alpha 5 |
| chr9_-_6661657 | 1.35 |
ENSDART00000133178
ENSDART00000113914 ENSDART00000061593 |
pou3f3a
|
POU class 3 homeobox 3a |
| chr16_-_35952789 | 1.33 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr9_+_53725294 | 1.33 |
ENSDART00000165991
|
cnmd
|
chondromodulin |
| chr9_+_53725128 | 1.33 |
ENSDART00000169062
|
cnmd
|
chondromodulin |
| chr9_+_44431174 | 1.33 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr21_+_43172506 | 1.33 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr6_+_41446541 | 1.31 |
ENSDART00000029553
ENSDART00000128756 ENSDART00000144864 |
rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr10_-_14545658 | 1.30 |
ENSDART00000057865
|
ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr18_-_44847855 | 1.30 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr18_-_19005919 | 1.29 |
ENSDART00000129776
|
ints14
|
integrator complex subunit 14 |
| chr21_+_30549512 | 1.28 |
ENSDART00000132831
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr1_-_55166511 | 1.26 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr2_-_32262287 | 1.25 |
ENSDART00000056621
ENSDART00000039717 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr23_-_44848961 | 1.25 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr19_-_10324182 | 1.23 |
ENSDART00000151352
ENSDART00000151162 ENSDART00000023571 |
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr20_+_29209767 | 1.22 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr2_+_25840463 | 1.22 |
ENSDART00000125178
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr23_+_36653376 | 1.22 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr16_+_20904754 | 1.20 |
ENSDART00000006043
|
hoxa11b
|
homeobox A11b |
| chr20_+_38671894 | 1.19 |
ENSDART00000146544
|
mpv17
|
MpV17 mitochondrial inner membrane protein |
| chr16_+_23961276 | 1.18 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr13_-_32726178 | 1.18 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr12_+_17154655 | 1.18 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr14_-_17588345 | 1.16 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr4_-_74466703 | 1.15 |
ENSDART00000174032
ENSDART00000189417 |
LO018124.1
|
|
| chr21_-_32081552 | 1.14 |
ENSDART00000135659
|
mat2b
|
methionine adenosyltransferase II, beta |
| chr8_-_16697912 | 1.13 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr21_-_40938382 | 1.13 |
ENSDART00000008593
|
yipf5
|
Yip1 domain family, member 5 |
| chr25_-_14433503 | 1.13 |
ENSDART00000103957
|
exoc3l1
|
exocyst complex component 3-like 1 |
| chr24_-_37272116 | 1.13 |
ENSDART00000022999
|
ube2ib
|
ubiquitin-conjugating enzyme E2Ib |
| chr18_-_40884087 | 1.13 |
ENSDART00000059194
|
snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr12_-_8504278 | 1.12 |
ENSDART00000135865
|
egr2b
|
early growth response 2b |
| chr1_+_26667872 | 1.11 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr2_-_44777592 | 1.09 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr21_-_26490186 | 1.09 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr16_-_30564319 | 1.08 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr17_+_51262556 | 1.08 |
ENSDART00000186748
ENSDART00000181606 ENSDART00000063738 ENSDART00000189066 |
eipr1
|
EARP complex and GARP complex interacting protein 1 |
| chr21_-_21020708 | 1.07 |
ENSDART00000064032
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr10_+_10728870 | 1.07 |
ENSDART00000109282
|
swi5
|
SWI5 homologous recombination repair protein |
| chr24_-_21172122 | 1.05 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr14_-_25040359 | 1.05 |
ENSDART00000162556
|
TMEM216
|
si:rp71-1d10.5 |
| chr6_-_13206255 | 1.05 |
ENSDART00000065373
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr17_-_42213285 | 1.04 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr16_-_44673851 | 1.04 |
ENSDART00000015139
|
dcaf13
|
ddb1 and cul4 associated factor 13 |
| chr5_+_49744713 | 1.03 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr5_+_26913120 | 1.03 |
ENSDART00000126609
|
tbx3b
|
T-box 3b |
| chr6_-_54444929 | 1.02 |
ENSDART00000154121
|
sys1
|
Sys1 golgi trafficking protein |
| chr21_-_17037907 | 1.02 |
ENSDART00000101263
|
ube2g1b
|
ubiquitin-conjugating enzyme E2G 1b (UBC7 homolog, yeast) |
| chr17_-_28707898 | 1.01 |
ENSDART00000135752
ENSDART00000061853 |
ap4s1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr3_+_61523035 | 1.01 |
ENSDART00000106530
|
znf1004
|
zinc finger protein 1004 |
| chr11_-_30634286 | 1.00 |
ENSDART00000191019
|
zgc:153665
|
zgc:153665 |
| chr21_-_39081107 | 1.00 |
ENSDART00000075935
|
vtnb
|
vitronectin b |
| chr23_-_16843649 | 0.99 |
ENSDART00000129971
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr7_+_26844261 | 0.99 |
ENSDART00000079165
|
ext2
|
exostosin glycosyltransferase 2 |
| chr3_-_37571601 | 0.99 |
ENSDART00000016407
|
arf2a
|
ADP-ribosylation factor 2a |
| chr16_+_23913943 | 0.99 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr21_+_26522571 | 0.99 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr15_+_21252532 | 0.98 |
ENSDART00000162619
ENSDART00000019636 ENSDART00000144901 ENSDART00000138676 ENSDART00000133821 ENSDART00000146967 ENSDART00000143990 ENSDART00000142070 ENSDART00000132373 |
usf1
|
upstream transcription factor 1 |
| chr7_+_38808027 | 0.98 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr5_-_63302944 | 0.97 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr22_+_20169352 | 0.97 |
ENSDART00000169055
ENSDART00000061617 |
hmg20b
|
high mobility group 20B |
| chr16_-_11986321 | 0.96 |
ENSDART00000148666
ENSDART00000029121 |
usp5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr4_+_42450386 | 0.96 |
ENSDART00000168211
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr14_-_31488100 | 0.95 |
ENSDART00000186246
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr20_+_30378803 | 0.95 |
ENSDART00000148242
ENSDART00000169140 ENSDART00000062441 |
rnaseh1
|
ribonuclease H1 |
| chr14_-_25985698 | 0.94 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr17_+_43595692 | 0.94 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr19_+_20178978 | 0.93 |
ENSDART00000145115
ENSDART00000151175 |
tra2a
|
transformer 2 alpha homolog |
| chr7_+_24023653 | 0.93 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr1_-_54622227 | 0.93 |
ENSDART00000049010
|
tekt4
|
tektin 4 |
| chr18_-_14337450 | 0.92 |
ENSDART00000061435
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr5_-_14509137 | 0.92 |
ENSDART00000180742
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr16_-_17713859 | 0.92 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr5_+_63302660 | 0.91 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr20_+_29209926 | 0.90 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr4_-_1824836 | 0.90 |
ENSDART00000111858
|
mrpl42
|
mitochondrial ribosomal protein L42 |
| chr4_-_12795030 | 0.90 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr16_+_33163858 | 0.90 |
ENSDART00000101943
|
rragca
|
Ras-related GTP binding Ca |
| chr24_-_25461267 | 0.89 |
ENSDART00000105820
|
mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr22_-_21176269 | 0.88 |
ENSDART00000112839
|
rex1bd
|
required for excision 1-B domain containing |
| chr11_-_31172276 | 0.86 |
ENSDART00000171520
|
si:dkey-238i5.3
|
si:dkey-238i5.3 |
| chr5_+_29726428 | 0.86 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr3_-_57762247 | 0.86 |
ENSDART00000156522
|
cant1a
|
calcium activated nucleotidase 1a |
| chr1_-_44940830 | 0.85 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr7_-_33023404 | 0.85 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr19_-_46018152 | 0.84 |
ENSDART00000159206
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr3_-_27065477 | 0.84 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr15_+_23528010 | 0.83 |
ENSDART00000152786
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr20_-_10120442 | 0.83 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr15_+_23528310 | 0.83 |
ENSDART00000152523
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr8_-_3312384 | 0.82 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr6_-_53048291 | 0.81 |
ENSDART00000103267
|
fam212ab
|
family with sequence similarity 212, member Ab |
| chr14_-_33894915 | 0.81 |
ENSDART00000143290
|
urp1
|
urotensin-related peptide 1 |
| chr17_+_8799661 | 0.80 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr18_+_6857071 | 0.79 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr8_-_20245892 | 0.79 |
ENSDART00000136911
|
acer1
|
alkaline ceramidase 1 |
| chr14_-_36799280 | 0.79 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr25_-_21280584 | 0.79 |
ENSDART00000143644
|
tmem60
|
transmembrane protein 60 |
| chr15_+_24644016 | 0.79 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr11_+_7528599 | 0.79 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr4_+_10721795 | 0.79 |
ENSDART00000136000
ENSDART00000067253 |
stab2
|
stabilin 2 |
| chr25_-_35101396 | 0.78 |
ENSDART00000138865
|
zgc:162611
|
zgc:162611 |
| chr23_+_27703749 | 0.77 |
ENSDART00000027224
|
lmbr1l
|
limb development membrane protein 1-like |
| chr6_-_40851499 | 0.77 |
ENSDART00000182591
|
rft1
|
RFT1 homolog |
| chr9_+_52411530 | 0.76 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr9_+_34950942 | 0.76 |
ENSDART00000077800
|
tfdp1a
|
transcription factor Dp-1, a |
| chr19_+_30867845 | 0.75 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr19_-_19720744 | 0.74 |
ENSDART00000170636
|
evx1
|
even-skipped homeobox 1 |
| chr15_-_7337537 | 0.74 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr6_+_13206516 | 0.74 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr25_-_35101673 | 0.74 |
ENSDART00000140864
|
zgc:162611
|
zgc:162611 |
| chr16_-_41714988 | 0.74 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr5_+_11944054 | 0.74 |
ENSDART00000159448
|
zgc:110063
|
zgc:110063 |
| chr23_+_9206350 | 0.73 |
ENSDART00000136236
|
si:dkey-66g10.2
|
si:dkey-66g10.2 |
| chr9_-_27719998 | 0.73 |
ENSDART00000161068
ENSDART00000148195 ENSDART00000138386 |
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr23_-_36449111 | 0.73 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr6_-_41135215 | 0.73 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr8_-_16712111 | 0.73 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr6_+_52895008 | 0.72 |
ENSDART00000136143
|
or137-7
|
odorant receptor, family H, subfamily 137, member 7 |
| chr6_+_52903302 | 0.71 |
ENSDART00000143962
|
or137-6
|
odorant receptor, family H, subfamily 137, member 6 |
| chr16_-_8280885 | 0.71 |
ENSDART00000129068
|
entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr7_-_37555208 | 0.71 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr11_+_2416064 | 0.70 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr14_-_29799993 | 0.69 |
ENSDART00000133775
ENSDART00000005568 |
pdlim3b
|
PDZ and LIM domain 3b |
| chr2_+_25839940 | 0.69 |
ENSDART00000139927
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr1_+_38362412 | 0.69 |
ENSDART00000075086
|
cep44
|
centrosomal protein 44 |
| chr2_-_32670883 | 0.69 |
ENSDART00000192586
ENSDART00000191410 |
puf60a
|
poly-U binding splicing factor a |
| chr25_-_25142387 | 0.69 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr8_+_49570884 | 0.69 |
ENSDART00000182117
ENSDART00000108613 |
rasef
|
RAS and EF-hand domain containing |
| chr3_+_32571929 | 0.69 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr2_-_37277626 | 0.68 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr6_-_10728057 | 0.67 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr21_+_40195452 | 0.66 |
ENSDART00000108650
|
or115-15
|
odorant receptor, family F, subfamily 115, member 15 |
| chr18_-_15532016 | 0.66 |
ENSDART00000165279
|
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr7_+_19850889 | 0.65 |
ENSDART00000100782
|
mus81
|
MUS81 structure-specific endonuclease subunit |
| chr13_+_42544009 | 0.65 |
ENSDART00000145409
|
si:dkey-221j11.3
|
si:dkey-221j11.3 |
| chr20_+_54274431 | 0.65 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr20_+_15015557 | 0.65 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr1_+_33647682 | 0.65 |
ENSDART00000114384
|
znf654
|
zinc finger protein 654 |
| chr4_-_20135919 | 0.64 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr11_-_7380674 | 0.64 |
ENSDART00000014979
ENSDART00000103418 |
vtg3
|
vitellogenin 3, phosvitinless |
| chr19_-_17996336 | 0.64 |
ENSDART00000186143
ENSDART00000080751 |
ints8
|
integrator complex subunit 8 |
| chr7_-_13906409 | 0.64 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr15_-_26552393 | 0.64 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr23_-_29357764 | 0.63 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr21_+_20386865 | 0.63 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr5_+_34622320 | 0.63 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr2_+_1988036 | 0.63 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr9_-_29497916 | 0.62 |
ENSDART00000060246
|
dnajc3a
|
DnaJ (Hsp40) homolog, subfamily C, member 3a |
| chr10_+_31951338 | 0.62 |
ENSDART00000019416
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr2_-_27330181 | 0.61 |
ENSDART00000137053
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr21_-_18991165 | 0.61 |
ENSDART00000012574
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr3_+_32492467 | 0.60 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr16_-_25233515 | 0.60 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr15_-_25571865 | 0.60 |
ENSDART00000077836
|
mmp20b
|
matrix metallopeptidase 20b (enamelysin) |
| chr15_+_7176182 | 0.60 |
ENSDART00000101578
|
her8.2
|
hairy-related 8.2 |
| chr7_-_27033080 | 0.59 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr8_-_14184423 | 0.59 |
ENSDART00000063817
|
ndufb11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxa3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.7 | 2.9 | GO:0006522 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.7 | 3.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.5 | 2.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.5 | 1.5 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.5 | 1.9 | GO:0016108 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.4 | 1.2 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.4 | 1.2 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.4 | 1.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 2.5 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.3 | 1.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 | 1.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 1.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.3 | 1.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 1.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 0.7 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.2 | 1.9 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 2.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 1.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 0.6 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.2 | 0.8 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 1.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 0.6 | GO:0046333 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.2 | 0.4 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 1.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.2 | 0.9 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 0.2 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 0.5 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 0.5 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.2 | 1.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 0.6 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 2.3 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.1 | 0.7 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 1.9 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 1.8 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.6 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 1.0 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.6 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 1.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.9 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.6 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 1.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.7 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 1.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.3 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 3.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.3 | GO:0034398 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 2.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.9 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 0.9 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 3.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 0.7 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 1.1 | GO:0048938 | myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.1 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.8 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.9 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 1.9 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 1.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 0.5 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) pharyngeal muscle development(GO:0043282) |
| 0.1 | 0.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.4 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 1.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.6 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 2.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 2.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.3 | GO:0090467 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.7 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 1.5 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.5 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.8 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 1.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.6 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 0.5 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 1.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 1.7 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.7 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 1.5 | GO:0003205 | cardiac chamber development(GO:0003205) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.5 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.2 | GO:0060052 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 1.0 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.5 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 1.9 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.4 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.6 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.6 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.8 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.0 | 0.6 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 1.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.5 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.5 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.4 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.9 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.4 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.5 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 1.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.1 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 1.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.0 | 0.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.6 | 2.3 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.4 | 1.9 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.3 | 1.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 1.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 3.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 1.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 0.9 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 0.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 0.5 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.4 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.7 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 1.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 1.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 1.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 1.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.7 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 1.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 1.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.2 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.1 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 0.3 | GO:0030132 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 3.3 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.5 | 1.9 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.5 | 1.8 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.4 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 1.9 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.3 | 1.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 1.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.2 | 1.0 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 1.0 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 0.6 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.2 | 0.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 2.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 1.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 1.6 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.6 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 3.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.7 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.1 | 0.5 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 1.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 3.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 2.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.2 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 1.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 1.0 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.2 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 5.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 2.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.7 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 3.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 4.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 0.9 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |