Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
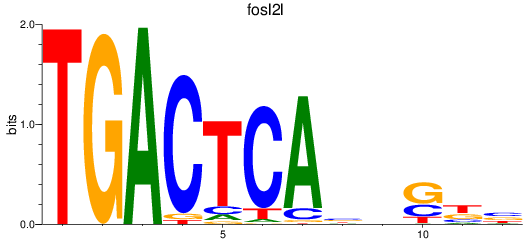
Results for fosl2l
Z-value: 0.80
Transcription factors associated with fosl2l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl2l
|
ENSDARG00000092174 | fos-like antigen 2 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-59d15.4 | dr11_v1_chr20_+_9211237_9211237 | 0.38 | 1.1e-01 | Click! |
Activity profile of fosl2l motif
Sorted Z-values of fosl2l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_40009446 | 2.36 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr2_-_293793 | 1.46 |
ENSDART00000082091
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr23_-_1017605 | 1.37 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr21_-_40880317 | 1.35 |
ENSDART00000100054
ENSDART00000137696 |
elnb
|
elastin b |
| chr15_-_47115787 | 1.19 |
ENSDART00000192601
|
CABZ01079081.1
|
|
| chr9_-_12493628 | 1.14 |
ENSDART00000180832
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr24_-_33366188 | 1.13 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr5_-_20678300 | 1.09 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr18_+_15841449 | 1.08 |
ENSDART00000141800
ENSDART00000091349 |
eea1
|
early endosome antigen 1 |
| chr23_-_1017428 | 1.08 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr19_-_21832441 | 0.93 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr13_-_298454 | 0.91 |
ENSDART00000135782
|
chs1
|
chitin synthase 1 |
| chr21_-_42100471 | 0.91 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr7_+_14005111 | 0.89 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr18_+_40355408 | 0.89 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr16_+_14033121 | 0.84 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr21_+_6290566 | 0.83 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr1_+_19708508 | 0.82 |
ENSDART00000054581
ENSDART00000131206 |
march1
|
membrane-associated ring finger (C3HC4) 1 |
| chr23_-_37113396 | 0.82 |
ENSDART00000102886
ENSDART00000134461 |
zgc:193690
|
zgc:193690 |
| chr14_-_970853 | 0.81 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr22_-_38543630 | 0.81 |
ENSDART00000172029
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr8_-_13211527 | 0.80 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr22_-_10110959 | 0.78 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr9_+_33154841 | 0.78 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr6_-_609880 | 0.78 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr22_+_19247255 | 0.76 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr6_-_40842768 | 0.76 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr12_+_46967789 | 0.75 |
ENSDART00000114866
|
oat
|
ornithine aminotransferase |
| chr19_+_21820144 | 0.74 |
ENSDART00000181996
|
CU856623.1
|
|
| chr23_-_45398622 | 0.74 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr8_+_6576940 | 0.72 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr19_+_4139065 | 0.72 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr17_-_3986236 | 0.71 |
ENSDART00000188794
ENSDART00000160830 |
PLCB1
|
si:ch1073-140o9.2 |
| chr2_-_155270 | 0.69 |
ENSDART00000131177
|
adcy1b
|
adenylate cyclase 1b |
| chr10_-_105100 | 0.69 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr21_+_43199237 | 0.68 |
ENSDART00000151748
|
aff4
|
AF4/FMR2 family, member 4 |
| chr10_-_20481763 | 0.67 |
ENSDART00000091156
ENSDART00000192358 |
tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr21_+_6328801 | 0.65 |
ENSDART00000163577
|
fnbp1b
|
formin binding protein 1b |
| chr7_-_29534001 | 0.63 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr16_+_49005321 | 0.62 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr8_+_3820134 | 0.62 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr22_+_36547670 | 0.62 |
ENSDART00000125991
ENSDART00000186372 |
armc9
|
armadillo repeat containing 9 |
| chr15_+_2559875 | 0.61 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr15_+_42235449 | 0.60 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr3_-_4303262 | 0.60 |
ENSDART00000112819
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr20_+_6543674 | 0.58 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr11_+_2596667 | 0.58 |
ENSDART00000175330
|
dnajc14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr9_+_2762270 | 0.58 |
ENSDART00000123342
ENSDART00000001795 ENSDART00000177563 |
sp3a
|
sp3a transcription factor |
| chr18_+_38900092 | 0.58 |
ENSDART00000148541
ENSDART00000133618 ENSDART00000088037 |
myo5aa
|
myosin VAa |
| chr2_-_3678029 | 0.57 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr17_-_43399896 | 0.57 |
ENSDART00000156033
ENSDART00000156418 |
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr11_-_10770053 | 0.57 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr9_+_46745348 | 0.56 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr23_-_31693309 | 0.55 |
ENSDART00000146327
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_-_34480599 | 0.54 |
ENSDART00000159602
ENSDART00000182248 |
si:dkeyp-51g9.4
si:ch211-64i20.3
|
si:dkeyp-51g9.4 si:ch211-64i20.3 |
| chr7_+_67486807 | 0.54 |
ENSDART00000159989
|
cpne7
|
copine VII |
| chr15_+_42767803 | 0.53 |
ENSDART00000164879
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr23_+_30967686 | 0.53 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr15_+_3766101 | 0.53 |
ENSDART00000042580
ENSDART00000112698 ENSDART00000187035 ENSDART00000165571 ENSDART00000121752 |
RNF14 (1 of many)
|
zmp:0000000524 |
| chr2_-_23768818 | 0.52 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr18_+_40354998 | 0.52 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr7_-_7764287 | 0.52 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr7_+_19482084 | 0.51 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr13_-_20381485 | 0.51 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr8_+_50190742 | 0.50 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr3_+_56905103 | 0.50 |
ENSDART00000124728
ENSDART00000154408 |
otop2
|
otopetrin 2 |
| chr3_+_24708685 | 0.50 |
ENSDART00000153507
|
mkl1a
|
megakaryoblastic leukemia (translocation) 1a |
| chr14_-_22100118 | 0.50 |
ENSDART00000157547
|
ssrp1a
|
structure specific recognition protein 1a |
| chr15_-_43978141 | 0.49 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr13_+_19283936 | 0.49 |
ENSDART00000111462
|
eif3s10
|
eukaryotic translation initiation factor 3, subunit 10 (theta) |
| chr15_+_1199407 | 0.49 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr4_-_47096170 | 0.49 |
ENSDART00000161079
|
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr6_+_42587637 | 0.48 |
ENSDART00000179964
|
camkva
|
CaM kinase-like vesicle-associated a |
| chr19_-_36675023 | 0.48 |
ENSDART00000132471
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr20_-_35578435 | 0.47 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr4_-_18775548 | 0.47 |
ENSDART00000141187
|
def6c
|
differentially expressed in FDCP 6c homolog |
| chr19_+_233143 | 0.47 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr5_-_13076779 | 0.47 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr4_-_1801519 | 0.46 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr19_-_7272921 | 0.46 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr10_+_31344227 | 0.45 |
ENSDART00000184470
|
AL845320.2
|
|
| chr16_+_48714048 | 0.45 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr21_-_4539899 | 0.45 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr11_+_6752468 | 0.45 |
ENSDART00000187046
ENSDART00000182089 |
pde4cb
|
phosphodiesterase 4C, cAMP-specific b |
| chr9_-_23181062 | 0.44 |
ENSDART00000192578
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr2_-_52020049 | 0.44 |
ENSDART00000128198
ENSDART00000135331 |
scn12aa
|
sodium channel, voltage gated, type XII, alpha a |
| chr3_+_4346854 | 0.44 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr23_-_36003441 | 0.44 |
ENSDART00000164699
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr8_+_14855471 | 0.44 |
ENSDART00000134775
|
si:dkey-21p1.3
|
si:dkey-21p1.3 |
| chr25_-_3549321 | 0.44 |
ENSDART00000181214
ENSDART00000160600 |
hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr10_-_22970723 | 0.43 |
ENSDART00000182145
ENSDART00000100802 |
FP102052.1
|
|
| chr9_+_30275139 | 0.43 |
ENSDART00000131781
|
rpgra
|
retinitis pigmentosa GTPase regulator a |
| chr18_+_16125852 | 0.43 |
ENSDART00000061106
|
bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr8_-_38403018 | 0.42 |
ENSDART00000134100
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr1_+_23230459 | 0.42 |
ENSDART00000147857
|
si:dkey-92j12.5
|
si:dkey-92j12.5 |
| chr12_+_1286642 | 0.42 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr8_+_10835456 | 0.42 |
ENSDART00000151388
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr9_-_34915351 | 0.42 |
ENSDART00000100728
ENSDART00000139608 |
upf3a
|
UPF3A, regulator of nonsense mediated mRNA decay |
| chr12_+_29054907 | 0.42 |
ENSDART00000152936
|
gabrz
|
gamma-aminobutyric acid (GABA) A receptor, zeta |
| chr4_+_58661331 | 0.41 |
ENSDART00000188763
|
CR388148.2
|
|
| chr12_+_18234557 | 0.41 |
ENSDART00000130741
|
fam20cb
|
family with sequence similarity 20, member Cb |
| chr24_-_23998897 | 0.41 |
ENSDART00000130053
|
zmp:0000000991
|
zmp:0000000991 |
| chr20_-_630637 | 0.41 |
ENSDART00000160454
|
si:dkey-121j17.5
|
si:dkey-121j17.5 |
| chr9_+_33158191 | 0.40 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr7_-_6458222 | 0.40 |
ENSDART00000172861
|
zgc:165555
|
zgc:165555 |
| chr8_+_48965767 | 0.39 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr22_+_26600834 | 0.39 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr5_+_61556172 | 0.39 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr6_-_33685325 | 0.39 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr23_+_19655301 | 0.38 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr4_+_40035116 | 0.38 |
ENSDART00000167808
|
BX649521.1
|
|
| chr25_-_31907590 | 0.38 |
ENSDART00000149707
ENSDART00000149471 |
eif3ja
|
eukaryotic translation initiation factor 3, subunit Ja |
| chr4_-_33073382 | 0.38 |
ENSDART00000146414
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr25_+_13498188 | 0.37 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr6_-_59357256 | 0.37 |
ENSDART00000074534
|
fam210b
|
family with sequence similarity 210, member B |
| chr22_-_6229275 | 0.37 |
ENSDART00000146045
ENSDART00000179730 |
si:ch211-274k16.2
|
si:ch211-274k16.2 |
| chr12_-_17147473 | 0.37 |
ENSDART00000106035
|
stambpl1
|
STAM binding protein-like 1 |
| chr21_+_13245302 | 0.37 |
ENSDART00000189498
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr18_+_618005 | 0.37 |
ENSDART00000189667
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr19_-_40776608 | 0.37 |
ENSDART00000179941
|
calcr
|
calcitonin receptor |
| chr8_+_52788997 | 0.36 |
ENSDART00000169198
|
adgrd2
|
adhesion G protein-coupled receptor D2 |
| chr15_+_32867420 | 0.36 |
ENSDART00000159442
|
dclk1b
|
doublecortin-like kinase 1b |
| chr4_-_75651505 | 0.35 |
ENSDART00000115143
|
si:dkey-71l4.2
|
si:dkey-71l4.2 |
| chr13_-_10945288 | 0.35 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr12_+_32292564 | 0.35 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr9_+_22388686 | 0.34 |
ENSDART00000182731
ENSDART00000181462 |
dgkg
|
diacylglycerol kinase, gamma |
| chr12_-_5455936 | 0.34 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr4_+_75575252 | 0.34 |
ENSDART00000166536
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr8_+_48966165 | 0.34 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr4_-_63558838 | 0.34 |
ENSDART00000191708
ENSDART00000190740 ENSDART00000186476 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr23_-_29003864 | 0.34 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr17_-_15600455 | 0.34 |
ENSDART00000110272
ENSDART00000156911 |
si:ch211-266g18.9
|
si:ch211-266g18.9 |
| chr1_+_10318089 | 0.34 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr8_-_11170114 | 0.33 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr6_+_18367388 | 0.33 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr7_+_39706004 | 0.33 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr2_-_13691834 | 0.32 |
ENSDART00000186570
|
CABZ01044235.1
|
|
| chr9_+_22466218 | 0.32 |
ENSDART00000131214
|
dgkg
|
diacylglycerol kinase, gamma |
| chr1_-_411331 | 0.32 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr10_+_40204175 | 0.32 |
ENSDART00000188277
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr21_+_3901775 | 0.32 |
ENSDART00000053609
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr24_+_38534550 | 0.32 |
ENSDART00000105677
|
zgc:154125
|
zgc:154125 |
| chr23_-_20010579 | 0.31 |
ENSDART00000089999
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr23_+_42254960 | 0.31 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr12_-_35787801 | 0.31 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr24_+_30215475 | 0.31 |
ENSDART00000164717
|
si:ch73-358j7.2
|
si:ch73-358j7.2 |
| chr18_+_25225524 | 0.31 |
ENSDART00000055563
|
si:dkeyp-59c12.1
|
si:dkeyp-59c12.1 |
| chr7_+_67381912 | 0.30 |
ENSDART00000167564
|
nfat5b
|
nuclear factor of activated T cells 5b |
| chr4_-_38829098 | 0.30 |
ENSDART00000151932
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr23_-_29004753 | 0.30 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr25_+_16117138 | 0.30 |
ENSDART00000146604
|
far1
|
fatty acyl CoA reductase 1 |
| chr4_+_65487457 | 0.29 |
ENSDART00000162017
ENSDART00000193241 |
si:dkeyp-5g9.1
|
si:dkeyp-5g9.1 |
| chr9_-_47654204 | 0.29 |
ENSDART00000015159
|
tns1b
|
tensin 1b |
| chr4_-_76303301 | 0.29 |
ENSDART00000158504
|
zgc:174944
|
zgc:174944 |
| chr4_+_68519945 | 0.28 |
ENSDART00000171148
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr5_-_22369916 | 0.28 |
ENSDART00000074964
|
si:dkey-27p18.2
|
si:dkey-27p18.2 |
| chr10_-_44482911 | 0.28 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr17_-_26911852 | 0.28 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr1_-_25177086 | 0.28 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr17_-_45247151 | 0.28 |
ENSDART00000186230
|
ttbk2a
|
tau tubulin kinase 2a |
| chr8_+_54274355 | 0.28 |
ENSDART00000067639
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr23_+_13814978 | 0.28 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr24_-_11325849 | 0.27 |
ENSDART00000182485
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr25_-_33275666 | 0.27 |
ENSDART00000193605
|
CABZ01046954.1
|
|
| chr17_-_45247332 | 0.27 |
ENSDART00000016815
|
ttbk2a
|
tau tubulin kinase 2a |
| chr3_+_15776446 | 0.27 |
ENSDART00000146651
|
znf652
|
zinc finger protein 652 |
| chr12_+_4573696 | 0.27 |
ENSDART00000152534
|
si:dkey-94f20.4
|
si:dkey-94f20.4 |
| chr4_-_38828508 | 0.26 |
ENSDART00000151952
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr21_+_20901505 | 0.26 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr6_+_40723554 | 0.26 |
ENSDART00000103833
|
slc26a6l
|
solute carrier family 26, member 6, like |
| chr4_+_45389591 | 0.26 |
ENSDART00000162530
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr1_-_7970653 | 0.26 |
ENSDART00000188909
|
si:dkey-79f11.7
|
si:dkey-79f11.7 |
| chr4_+_34121902 | 0.26 |
ENSDART00000170225
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr4_+_58667348 | 0.26 |
ENSDART00000186596
ENSDART00000180673 |
CR388148.2
|
|
| chr23_+_19759128 | 0.26 |
ENSDART00000135820
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr4_-_32646394 | 0.26 |
ENSDART00000143196
|
si:dkey-265e7.1
|
si:dkey-265e7.1 |
| chr24_+_42074143 | 0.26 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr15_-_23443588 | 0.26 |
ENSDART00000170427
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr4_-_73787702 | 0.25 |
ENSDART00000136328
ENSDART00000150546 |
si:dkey-262g12.3
|
si:dkey-262g12.3 |
| chr13_-_37545487 | 0.25 |
ENSDART00000131269
|
syt16
|
synaptotagmin XVI |
| chr23_-_7799184 | 0.25 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr12_+_47280839 | 0.25 |
ENSDART00000187574
|
RYR2
|
ryanodine receptor 2 |
| chr23_-_11128601 | 0.25 |
ENSDART00000131232
|
cntn3a.2
|
contactin 3a, tandem duplicate 2 |
| chr8_+_5024468 | 0.25 |
ENSDART00000030938
|
adra1aa
|
adrenoceptor alpha 1Aa |
| chr4_-_39487825 | 0.24 |
ENSDART00000161855
|
si:dkey-261o4.6
|
si:dkey-261o4.6 |
| chr23_-_37113215 | 0.24 |
ENSDART00000146835
|
zgc:193690
|
zgc:193690 |
| chr23_-_7797207 | 0.24 |
ENSDART00000181611
|
myt1b
|
myelin transcription factor 1b |
| chr4_+_35227135 | 0.24 |
ENSDART00000161023
ENSDART00000182403 |
si:ch211-276i6.1
|
si:ch211-276i6.1 |
| chr11_+_37905630 | 0.24 |
ENSDART00000170303
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr4_-_52783184 | 0.24 |
ENSDART00000172283
|
si:dkey-4j21.2
|
si:dkey-4j21.2 |
| chr18_+_3052408 | 0.23 |
ENSDART00000181563
|
kctd14
|
potassium channel tetramerization domain containing 14 |
| chr23_+_9067131 | 0.23 |
ENSDART00000144533
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr18_-_8396192 | 0.23 |
ENSDART00000193830
|
BEND7
|
si:ch211-220f12.4 |
| chr23_+_4414343 | 0.23 |
ENSDART00000081821
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr4_-_67984478 | 0.23 |
ENSDART00000160623
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr4_-_76105527 | 0.23 |
ENSDART00000159108
|
zgc:110171
|
zgc:110171 |
| chr15_-_42736433 | 0.23 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr23_+_4414164 | 0.22 |
ENSDART00000192762
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr20_+_23501535 | 0.22 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr3_-_48259289 | 0.22 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosl2l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 0.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 0.6 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.2 | 0.9 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.2 | 0.9 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 0.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.6 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.4 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.1 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.8 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.5 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.7 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.9 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.6 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.3 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 1.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.3 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.0 | 0.4 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 1.4 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.2 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 1.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.5 | GO:0007164 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 0.6 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.8 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.5 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 2.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.9 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 2.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.4 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.6 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.4 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.9 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 0.5 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 1.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.4 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.8 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.2 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |