Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
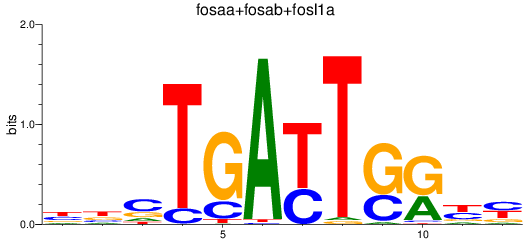
Results for fosaa+fosab+fosl1a
Z-value: 3.89
Transcription factors associated with fosaa+fosab+fosl1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl1a
|
ENSDARG00000015355 | FOS-like antigen 1a |
|
fosab
|
ENSDARG00000031683 | v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
|
fosaa
|
ENSDARG00000040135 | v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| fosaa | dr11_v1_chr17_-_50234004_50234004 | -0.80 | 3.6e-05 | Click! |
| fosab | dr11_v1_chr20_-_46554440_46554440 | -0.75 | 2.0e-04 | Click! |
| fosl1a | dr11_v1_chr14_-_30747686_30747686 | -0.24 | 3.2e-01 | Click! |
Activity profile of fosaa+fosab+fosl1a motif
Sorted Z-values of fosaa+fosab+fosl1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_16941319 | 16.53 |
ENSDART00000109968
|
zgc:174855
|
zgc:174855 |
| chr12_-_16694092 | 15.39 |
ENSDART00000047916
|
ctslb
|
cathepsin Lb |
| chr12_-_16877136 | 15.16 |
ENSDART00000152593
|
si:dkey-269i1.4
|
si:dkey-269i1.4 |
| chr12_-_16636627 | 14.10 |
ENSDART00000128811
|
si:dkey-239j18.3
|
si:dkey-239j18.3 |
| chr12_-_16619757 | 14.05 |
ENSDART00000145570
|
ctslb
|
cathepsin Lb |
| chr12_-_16558106 | 13.98 |
ENSDART00000109033
|
si:dkey-269i1.4
|
si:dkey-269i1.4 |
| chr12_-_16923162 | 13.33 |
ENSDART00000106072
|
si:dkey-26g8.5
|
si:dkey-26g8.5 |
| chr12_+_16949391 | 12.56 |
ENSDART00000152635
|
zgc:174153
|
zgc:174153 |
| chr12_-_16898140 | 11.50 |
ENSDART00000152656
|
MGC174155
|
Cathepsin L1-like |
| chr12_-_16619449 | 11.41 |
ENSDART00000182074
|
ctslb
|
cathepsin Lb |
| chr6_-_50704689 | 11.11 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr12_-_16720196 | 10.76 |
ENSDART00000187639
|
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr12_-_16720432 | 9.30 |
ENSDART00000152261
ENSDART00000152154 |
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr12_-_16595177 | 9.09 |
ENSDART00000133962
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr12_-_16595406 | 9.01 |
ENSDART00000166798
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr3_-_25377163 | 8.58 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr1_+_50921266 | 8.58 |
ENSDART00000006538
|
otx2a
|
orthodenticle homeobox 2a |
| chr21_-_5077715 | 8.50 |
ENSDART00000081954
|
haus1
|
HAUS augmin-like complex, subunit 1 |
| chr20_-_2641233 | 8.03 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr5_-_54712159 | 7.70 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr23_-_45705525 | 7.68 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr21_+_20715020 | 7.65 |
ENSDART00000015224
|
gadd45gb.1
|
growth arrest and DNA-damage-inducible, gamma b, tandem duplicate 1 |
| chr25_-_7520937 | 7.39 |
ENSDART00000170050
|
cdkn1cb
|
cyclin-dependent kinase inhibitor 1Cb |
| chr6_-_55297274 | 7.17 |
ENSDART00000184283
|
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr8_+_16738282 | 6.97 |
ENSDART00000134265
ENSDART00000100698 |
ercc8
|
excision repair cross-complementation group 8 |
| chr10_+_39283985 | 6.57 |
ENSDART00000016464
|
dcps
|
decapping enzyme, scavenger |
| chr2_-_17115256 | 6.52 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr7_+_22981909 | 6.41 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr7_+_22982201 | 6.36 |
ENSDART00000134116
|
ccnb3
|
cyclin B3 |
| chr1_-_33647138 | 6.35 |
ENSDART00000142111
ENSDART00000015547 |
cldng
|
claudin g |
| chr1_+_30723677 | 6.32 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr3_+_12554801 | 6.29 |
ENSDART00000167177
|
ccnf
|
cyclin F |
| chr23_+_44634187 | 6.29 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr13_+_46941930 | 6.26 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr2_-_47431205 | 6.26 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr17_+_19481049 | 6.12 |
ENSDART00000024194
|
kif11
|
kinesin family member 11 |
| chr11_+_29537756 | 6.05 |
ENSDART00000103388
|
wu:fi42e03
|
wu:fi42e03 |
| chr12_+_10706772 | 6.03 |
ENSDART00000158227
|
top2a
|
DNA topoisomerase II alpha |
| chr17_+_20923691 | 6.02 |
ENSDART00000122407
|
cdk1
|
cyclin-dependent kinase 1 |
| chr7_-_17337233 | 5.90 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr1_+_30723380 | 5.88 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr20_+_36623807 | 5.79 |
ENSDART00000149171
ENSDART00000062895 |
srp9
|
signal recognition particle 9 |
| chr5_-_14500622 | 5.75 |
ENSDART00000099566
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr14_+_21754521 | 5.68 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr9_+_15890558 | 5.57 |
ENSDART00000144032
|
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr4_+_4803698 | 5.49 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr5_+_13647288 | 5.48 |
ENSDART00000099660
ENSDART00000139199 |
h2afva
|
H2A histone family, member Va |
| chr11_+_14321113 | 5.46 |
ENSDART00000039822
ENSDART00000137347 ENSDART00000132997 |
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr5_+_1493767 | 5.36 |
ENSDART00000022132
|
haus4
|
HAUS augmin-like complex, subunit 4 |
| chr15_-_4528326 | 5.33 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr5_+_26913120 | 5.31 |
ENSDART00000126609
|
tbx3b
|
T-box 3b |
| chr11_-_36350421 | 5.28 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr7_+_30626378 | 5.26 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr14_-_26704829 | 5.24 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr10_+_10728870 | 5.24 |
ENSDART00000109282
|
swi5
|
SWI5 homologous recombination repair protein |
| chr23_-_21471022 | 5.22 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr11_-_10456553 | 5.17 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr16_+_40575742 | 5.11 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr19_+_15443353 | 5.08 |
ENSDART00000135923
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr20_-_36416922 | 5.07 |
ENSDART00000019145
|
lbr
|
lamin B receptor |
| chr10_-_35257458 | 5.05 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr1_-_33557915 | 5.01 |
ENSDART00000075632
|
creb1a
|
cAMP responsive element binding protein 1a |
| chr19_-_47782916 | 5.01 |
ENSDART00000063337
|
cdca8
|
cell division cycle associated 8 |
| chr11_-_10456387 | 4.96 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr21_+_45316330 | 4.89 |
ENSDART00000056474
ENSDART00000149314 ENSDART00000149272 ENSDART00000149156 ENSDART00000099497 |
tcf7
|
transcription factor 7 |
| chr10_+_16036573 | 4.86 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr5_+_43870389 | 4.80 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr4_-_2545310 | 4.70 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr19_-_47782586 | 4.68 |
ENSDART00000177126
|
cdca8
|
cell division cycle associated 8 |
| chr9_+_13120419 | 4.64 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr3_-_15999501 | 4.64 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr22_+_23546926 | 4.62 |
ENSDART00000157940
|
aspm
|
abnormal spindle microtubule assembly |
| chr1_-_54972170 | 4.61 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr1_+_58977262 | 4.61 |
ENSDART00000168832
|
eri2
|
ERI1 exoribonuclease family member 2 |
| chr4_-_16876281 | 4.57 |
ENSDART00000016690
ENSDART00000044005 ENSDART00000042874 ENSDART00000125762 ENSDART00000185974 |
tmpoa
|
thymopoietin a |
| chr13_+_2394264 | 4.57 |
ENSDART00000168595
|
elovl5
|
ELOVL fatty acid elongase 5 |
| chr2_-_17114852 | 4.56 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr12_-_17655683 | 4.55 |
ENSDART00000066411
|
dlgap5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr1_-_26071535 | 4.54 |
ENSDART00000185187
|
pdcd4a
|
programmed cell death 4a |
| chr20_-_29498178 | 4.53 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr10_+_16036246 | 4.53 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr5_+_41510387 | 4.47 |
ENSDART00000023779
|
vcp
|
valosin containing protein |
| chr3_-_24681404 | 4.47 |
ENSDART00000161612
|
BX569789.1
|
|
| chr10_-_10864331 | 4.47 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr7_+_57088920 | 4.45 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr2_+_26288301 | 4.44 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr25_+_37446861 | 4.41 |
ENSDART00000189250
|
FO834799.1
|
|
| chr23_+_21473103 | 4.39 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr2_+_105748 | 4.33 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr12_-_10674606 | 4.31 |
ENSDART00000157919
|
med24
|
mediator complex subunit 24 |
| chr18_+_62932 | 4.28 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr15_-_44052927 | 4.26 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr12_+_25945560 | 4.25 |
ENSDART00000109799
|
mmrn2b
|
multimerin 2b |
| chr11_-_36350876 | 4.23 |
ENSDART00000146495
ENSDART00000020655 |
psma5
|
proteasome subunit alpha 5 |
| chr10_+_32058692 | 4.23 |
ENSDART00000062309
|
thap12a
|
THAP domain containing 12a |
| chr9_+_32178374 | 4.22 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr14_-_970853 | 4.18 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr7_-_33684632 | 4.17 |
ENSDART00000130553
|
tle3b
|
transducin-like enhancer of split 3b |
| chr25_+_14507567 | 4.14 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr1_+_23563691 | 4.13 |
ENSDART00000142879
|
ncapg
|
non-SMC condensin I complex, subunit G |
| chr1_+_43686251 | 4.11 |
ENSDART00000074604
ENSDART00000137791 |
cisd2
|
CDGSH iron sulfur domain 2 |
| chr7_+_5965611 | 4.10 |
ENSDART00000115062
|
CU459186.2
|
Histone H3.2 |
| chr12_+_23424108 | 4.05 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr17_+_25563979 | 4.03 |
ENSDART00000045615
ENSDART00000183162 |
qrsl1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr19_+_6990970 | 4.03 |
ENSDART00000158758
ENSDART00000160482 ENSDART00000193566 |
kifc1
|
kinesin family member C1 |
| chr13_-_33256667 | 4.03 |
ENSDART00000003314
|
nusap1
|
nucleolar and spindle associated protein 1 |
| chr3_-_58644920 | 4.02 |
ENSDART00000155953
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr7_-_22981796 | 4.01 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr25_-_32363341 | 3.99 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr8_-_2591654 | 3.98 |
ENSDART00000049109
|
seta
|
SET nuclear proto-oncogene a |
| chr8_+_49766338 | 3.95 |
ENSDART00000060657
|
rmi1
|
RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) |
| chr7_+_29115890 | 3.94 |
ENSDART00000052345
|
tradd
|
tnfrsf1a-associated via death domain |
| chr16_-_12060488 | 3.93 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr25_-_37191929 | 3.91 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr12_+_13344896 | 3.90 |
ENSDART00000089017
|
rnasen
|
ribonuclease type III, nuclear |
| chr22_+_15960005 | 3.90 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr16_-_25680666 | 3.90 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr23_-_3759692 | 3.89 |
ENSDART00000028885
|
hmga1a
|
high mobility group AT-hook 1a |
| chr11_-_40519886 | 3.87 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr4_+_13428993 | 3.85 |
ENSDART00000067151
|
si:dkey-39a18.1
|
si:dkey-39a18.1 |
| chr25_-_6389713 | 3.85 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr13_-_25198025 | 3.85 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
| chr19_+_15443540 | 3.80 |
ENSDART00000193355
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr1_+_227241 | 3.79 |
ENSDART00000003317
|
tfdp1b
|
transcription factor Dp-1, b |
| chr2_+_33926911 | 3.78 |
ENSDART00000109849
ENSDART00000135884 |
kif2c
|
kinesin family member 2C |
| chr6_-_9676108 | 3.77 |
ENSDART00000169915
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr17_+_16423721 | 3.77 |
ENSDART00000064233
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr3_+_15893039 | 3.73 |
ENSDART00000055780
|
jpt2
|
Jupiter microtubule associated homolog 2 |
| chr23_-_3759345 | 3.73 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr20_-_53996193 | 3.72 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr14_-_46374870 | 3.71 |
ENSDART00000185803
ENSDART00000188313 ENSDART00000031498 |
ccna2
|
cyclin A2 |
| chr1_+_49352900 | 3.68 |
ENSDART00000008468
|
msx1b
|
muscle segment homeobox 1b |
| chr25_+_19095231 | 3.68 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr22_-_4439311 | 3.68 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr2_-_39675829 | 3.67 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr16_-_12060770 | 3.66 |
ENSDART00000183237
ENSDART00000103948 |
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr8_-_11229523 | 3.66 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr5_+_32791245 | 3.65 |
ENSDART00000077189
|
ier5l
|
immediate early response 5-like |
| chr9_+_4675193 | 3.65 |
ENSDART00000081326
|
prpf40a
|
PRP40 pre-mRNA processing factor 40 homolog A |
| chr15_-_43287515 | 3.64 |
ENSDART00000155103
|
prss16
|
protease, serine, 16 (thymus) |
| chr2_+_33926606 | 3.63 |
ENSDART00000111430
|
kif2c
|
kinesin family member 2C |
| chr22_+_15959844 | 3.63 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr9_-_40683722 | 3.62 |
ENSDART00000141979
ENSDART00000181228 |
bard1
|
BRCA1 associated RING domain 1 |
| chr9_+_29548195 | 3.60 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr16_+_40043673 | 3.60 |
ENSDART00000102552
ENSDART00000125484 |
trmt11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr25_+_7435291 | 3.60 |
ENSDART00000172567
ENSDART00000163017 |
prc1a
|
protein regulator of cytokinesis 1a |
| chr15_-_4596623 | 3.59 |
ENSDART00000132227
|
eif4h
|
eukaryotic translation initiation factor 4h |
| chr15_+_46329149 | 3.58 |
ENSDART00000128404
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr5_-_8171625 | 3.57 |
ENSDART00000167643
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr13_+_24662238 | 3.56 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr8_-_51753604 | 3.56 |
ENSDART00000007090
|
tbx16
|
T-box 16 |
| chr4_+_42450386 | 3.54 |
ENSDART00000168211
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr14_-_32744464 | 3.51 |
ENSDART00000075617
|
sox3
|
SRY (sex determining region Y)-box 3 |
| chr21_+_19070921 | 3.49 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr4_-_11603149 | 3.49 |
ENSDART00000150468
|
net1
|
neuroepithelial cell transforming 1 |
| chr8_+_26868105 | 3.48 |
ENSDART00000005337
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr18_-_25051846 | 3.48 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr19_-_35450857 | 3.47 |
ENSDART00000179357
|
anln
|
anillin, actin binding protein |
| chr25_-_6261693 | 3.45 |
ENSDART00000135808
|
ireb2
|
iron-responsive element binding protein 2 |
| chr6_+_269204 | 3.45 |
ENSDART00000191678
|
atf4a
|
activating transcription factor 4a |
| chr22_-_24984733 | 3.44 |
ENSDART00000142147
ENSDART00000187284 |
dnal4b
|
dynein, axonemal, light chain 4b |
| chr19_-_35450661 | 3.44 |
ENSDART00000113574
ENSDART00000136895 |
anln
|
anillin, actin binding protein |
| chr16_-_13680692 | 3.43 |
ENSDART00000047452
|
ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr19_-_2231146 | 3.43 |
ENSDART00000181909
ENSDART00000043595 |
twist1a
|
twist family bHLH transcription factor 1a |
| chr20_-_25626198 | 3.41 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr19_-_24125457 | 3.40 |
ENSDART00000080632
|
zgc:64022
|
zgc:64022 |
| chr14_-_246342 | 3.40 |
ENSDART00000054823
|
aurkb
|
aurora kinase B |
| chr23_-_4704938 | 3.39 |
ENSDART00000067293
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr8_-_52715911 | 3.37 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr21_-_14826066 | 3.37 |
ENSDART00000067001
|
noc4l
|
nucleolar complex associated 4 homolog |
| chr12_+_47663419 | 3.35 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr6_+_26948093 | 3.34 |
ENSDART00000153595
|
farp2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr1_-_12394048 | 3.33 |
ENSDART00000146067
ENSDART00000134708 |
sclt1
|
sodium channel and clathrin linker 1 |
| chr23_+_21459263 | 3.33 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr1_-_30723241 | 3.32 |
ENSDART00000152175
ENSDART00000152150 |
MZT1
|
si:dkey-15d12.2 |
| chr7_-_18547420 | 3.31 |
ENSDART00000173969
|
rgs12a
|
regulator of G protein signaling 12a |
| chr20_+_34770197 | 3.30 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr9_-_10068004 | 3.30 |
ENSDART00000011922
ENSDART00000162818 |
spopla
|
speckle-type POZ protein-like a |
| chr13_-_42536642 | 3.29 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr10_+_29816681 | 3.29 |
ENSDART00000100022
|
h2afx1
|
H2A histone family member X1 |
| chr21_-_929293 | 3.28 |
ENSDART00000006419
|
txnl1
|
thioredoxin-like 1 |
| chr7_-_33350082 | 3.28 |
ENSDART00000008785
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr5_-_54714789 | 3.28 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr12_-_35885349 | 3.27 |
ENSDART00000085662
|
cep131
|
centrosomal protein 131 |
| chr12_-_49151326 | 3.26 |
ENSDART00000153244
|
bub3
|
BUB3 mitotic checkpoint protein |
| chr13_+_12671513 | 3.25 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr1_-_54971968 | 3.24 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr11_-_42099645 | 3.24 |
ENSDART00000173312
|
abhd6a
|
abhydrolase domain containing 6a |
| chr5_+_69785990 | 3.23 |
ENSDART00000162057
ENSDART00000166893 |
kmt5ab
|
lysine methyltransferase 5Ab |
| chr13_+_36585399 | 3.23 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr7_+_22981441 | 3.22 |
ENSDART00000182887
|
ccnb3
|
cyclin B3 |
| chr12_+_5048044 | 3.22 |
ENSDART00000161548
ENSDART00000172607 |
kif22
|
kinesin family member 22 |
| chr5_-_36948586 | 3.20 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr23_-_33775145 | 3.19 |
ENSDART00000132147
ENSDART00000027959 ENSDART00000160116 |
racgap1
|
Rac GTPase activating protein 1 |
| chr25_+_28776562 | 3.18 |
ENSDART00000109702
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
| chr25_+_36292057 | 3.14 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr6_+_56147812 | 3.14 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr8_+_23639124 | 3.14 |
ENSDART00000083108
|
nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr19_+_14158075 | 3.14 |
ENSDART00000170335
ENSDART00000168260 |
nudc
|
nudC nuclear distribution protein |
| chr17_-_10000339 | 3.12 |
ENSDART00000162893
|
snx6
|
sorting nexin 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosaa+fosab+fosl1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 4.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 1.7 | 6.9 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 1.6 | 6.3 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 1.5 | 13.9 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 1.5 | 4.5 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 1.5 | 4.5 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 1.4 | 10.0 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 1.4 | 7.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.4 | 9.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 1.3 | 2.5 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 1.3 | 6.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.2 | 15.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 1.2 | 4.9 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 1.2 | 3.6 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 1.2 | 2.4 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 1.2 | 3.6 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) |
| 1.2 | 5.8 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 1.1 | 8.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.1 | 10.3 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.1 | 5.7 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 1.1 | 3.3 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 1.1 | 4.4 | GO:0060471 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 1.1 | 7.5 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 1.1 | 8.4 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 1.0 | 5.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 1.0 | 4.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 1.0 | 7.1 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 1.0 | 7.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 1.0 | 10.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 1.0 | 2.9 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.9 | 4.7 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.9 | 1.9 | GO:0009139 | pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) |
| 0.9 | 6.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.9 | 3.7 | GO:0045429 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.9 | 11.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.9 | 4.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.9 | 5.4 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.9 | 4.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.8 | 4.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.8 | 4.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.8 | 2.5 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.8 | 2.4 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.8 | 4.8 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.8 | 2.4 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.8 | 3.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.8 | 3.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.8 | 6.1 | GO:0090317 | negative regulation of intracellular protein transport(GO:0090317) |
| 0.8 | 30.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.7 | 6.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.7 | 8.9 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.7 | 4.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.7 | 5.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.7 | 2.9 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.7 | 4.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.7 | 8.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.7 | 4.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.7 | 2.1 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.7 | 6.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.7 | 7.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.7 | 2.1 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.7 | 2.0 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.7 | 8.7 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.7 | 3.3 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.6 | 9.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.6 | 14.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.6 | 8.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.6 | 2.5 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.6 | 3.1 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.6 | 3.7 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.6 | 15.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.6 | 1.8 | GO:0000052 | citrulline metabolic process(GO:0000052) citrulline biosynthetic process(GO:0019240) |
| 0.6 | 7.0 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.6 | 4.6 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.6 | 4.6 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.6 | 3.4 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.6 | 1.7 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.6 | 4.5 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.6 | 2.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.6 | 2.2 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.6 | 1.7 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.5 | 2.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.5 | 1.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.5 | 1.6 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.5 | 1.6 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.5 | 3.6 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.5 | 1.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.5 | 1.5 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.5 | 2.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.5 | 10.2 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.5 | 1.9 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.5 | 4.8 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.5 | 2.9 | GO:0007100 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.5 | 3.3 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.5 | 8.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.5 | 15.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.5 | 4.6 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.5 | 8.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.5 | 1.8 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.5 | 5.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.5 | 4.6 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.5 | 1.8 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.5 | 4.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.5 | 4.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.4 | 2.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.4 | 7.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.4 | 1.8 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.4 | 4.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.4 | 13.9 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.4 | 2.2 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.4 | 2.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.4 | 3.7 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.4 | 4.5 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.4 | 1.6 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.4 | 1.6 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.4 | 1.6 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.4 | 1.6 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.4 | 2.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.4 | 1.1 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.4 | 3.0 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.4 | 5.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.4 | 2.2 | GO:0003151 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.4 | 3.3 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.4 | 2.9 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.4 | 1.4 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.4 | 9.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.3 | 2.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 1.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 14.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.3 | 1.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.3 | 4.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.3 | 2.3 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.3 | 3.3 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.3 | 1.3 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.3 | 2.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 2.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.3 | 3.8 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.3 | 1.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.3 | 1.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.3 | 4.3 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.3 | 1.2 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.3 | 3.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.3 | 0.9 | GO:0060623 | regulation of sister chromatid cohesion(GO:0007063) regulation of chromosome condensation(GO:0060623) regulation of cohesin loading(GO:0071922) |
| 0.3 | 3.5 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.3 | 2.0 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.3 | 15.9 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.3 | 1.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.3 | 1.9 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.3 | 1.1 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.3 | 2.9 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 2.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 2.1 | GO:0048897 | myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.3 | 1.0 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.3 | 2.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.3 | 5.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.3 | 3.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 1.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 2.0 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) |
| 0.2 | 0.7 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 1.0 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 1.5 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 2.6 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) mitotic DNA integrity checkpoint(GO:0044774) |
| 0.2 | 1.7 | GO:0046323 | glucose import(GO:0046323) |
| 0.2 | 1.4 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.2 | 5.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.2 | 8.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 5.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 3.9 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.2 | 1.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.1 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.2 | 1.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 1.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.2 | 0.9 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.2 | 6.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 0.9 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 1.3 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.2 | 3.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 2.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 0.9 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.2 | 1.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 4.9 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 2.7 | GO:0034333 | adherens junction assembly(GO:0034333) |
| 0.2 | 1.7 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 0.6 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.2 | 2.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 1.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.2 | 1.2 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.2 | 6.3 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.2 | 4.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 4.0 | GO:0030168 | platelet activation(GO:0030168) |
| 0.2 | 1.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 0.6 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 1.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 2.6 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.2 | 3.6 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.2 | 0.6 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.2 | 1.7 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.2 | 2.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 3.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 7.3 | GO:0003309 | type B pancreatic cell differentiation(GO:0003309) |
| 0.2 | 0.9 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 2.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 1.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 2.5 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.2 | 4.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 3.7 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.2 | 1.4 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 1.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 116.1 | GO:0006955 | immune response(GO:0006955) |
| 0.2 | 2.7 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.2 | 1.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 3.4 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.2 | 5.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.2 | 0.5 | GO:0072388 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.2 | 2.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 9.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.2 | 1.5 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.2 | 1.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.9 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 3.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 1.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 0.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 0.9 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.2 | 1.4 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 2.4 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 1.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 7.8 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 0.7 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 0.7 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 10.8 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.1 | 1.0 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.2 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.1 | 1.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.4 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 1.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.7 | GO:1902623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of chemotaxis(GO:0050922) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.5 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 0.7 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.1 | 6.4 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 2.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 1.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 3.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 2.7 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 1.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 2.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 5.3 | GO:0003205 | cardiac chamber development(GO:0003205) |
| 0.1 | 1.0 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 2.4 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 1.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.6 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 2.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.7 | GO:1903038 | negative regulation of homotypic cell-cell adhesion(GO:0034111) negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.1 | 3.7 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 0.8 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 4.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 2.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.7 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 2.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 1.1 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.1 | 7.9 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 | 0.9 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.3 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.9 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 3.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.7 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.1 | 1.4 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 2.4 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 4.8 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.1 | 1.7 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 1.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 3.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 5.4 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 1.9 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 5.9 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 1.2 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 3.0 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 0.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.7 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 1.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.7 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.6 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 0.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 1.8 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 0.4 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 3.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.5 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 1.6 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.1 | 6.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 1.1 | GO:0031935 | methylation-dependent chromatin silencing(GO:0006346) regulation of chromatin silencing(GO:0031935) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.3 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.1 | 0.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 1.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.9 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.5 | GO:0007051 | spindle organization(GO:0007051) |
| 0.1 | 1.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 1.6 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 1.8 | GO:0060840 | artery development(GO:0060840) |
| 0.1 | 0.7 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 2.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 0.7 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.9 | GO:1903725 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) regulation of phospholipid metabolic process(GO:1903725) |
| 0.1 | 1.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.6 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 2.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 2.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 4.8 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.1 | 0.8 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.1 | 0.5 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.1 | 0.3 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 1.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 3.6 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 1.6 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 0.5 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 7.2 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.1 | 3.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.8 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 10.8 | GO:0032259 | methylation(GO:0032259) |
| 0.1 | 0.9 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.1 | 3.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 2.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.9 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.1 | 1.1 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 2.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 9.1 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 1.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 1.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 4.5 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.0 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 14.0 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 1.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 3.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0043901 | negative regulation of multi-organism process(GO:0043901) |
| 0.0 | 0.8 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 1.9 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 0.3 | GO:0042438 | melanin biosynthetic process(GO:0042438) secondary metabolite biosynthetic process(GO:0044550) |
| 0.0 | 1.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.6 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 1.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.5 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 2.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.4 | GO:0048515 | spermatid development(GO:0007286) spermatid differentiation(GO:0048515) |
| 0.0 | 3.0 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 1.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.0 | 1.3 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.0 | 0.3 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.4 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.6 | GO:0030335 | positive regulation of cell migration(GO:0030335) |
| 0.0 | 1.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.5 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 1.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 1.8 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 0.3 | GO:1901184 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) regulation of ERBB signaling pathway(GO:1901184) |
| 0.0 | 0.8 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.9 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.2 | GO:0060026 | convergent extension(GO:0060026) |
| 0.0 | 0.1 | GO:0033499 | hexose catabolic process(GO:0019320) galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 1.7 | GO:0045165 | cell fate commitment(GO:0045165) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 2.6 | GO:0006974 | cellular response to DNA damage stimulus(GO:0006974) |
| 0.0 | 1.2 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 0.1 | GO:0050658 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 | 0.2 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.3 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.6 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 5.6 | GO:0051726 | regulation of cell cycle(GO:0051726) |
| 0.0 | 0.5 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 2.6 | GO:0006412 | translation(GO:0006412) peptide biosynthetic process(GO:0043043) |
| 0.0 | 0.1 | GO:0061550 | cranial ganglion development(GO:0061550) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.9 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 2.6 | 15.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 2.0 | 6.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.8 | 16.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 1.8 | 5.4 | GO:0098536 | deuterosome(GO:0098536) |
| 1.6 | 1.6 | GO:0034709 | methylosome(GO:0034709) pICln-Sm protein complex(GO:0034715) |
| 1.3 | 5.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 1.2 | 3.7 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 1.2 | 13.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.1 | 3.4 | GO:0030689 | Noc complex(GO:0030689) |
| 1.1 | 4.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.0 | 7.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.0 | 7.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.9 | 32.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.8 | 5.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.8 | 5.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.8 | 10.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.8 | 4.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.7 | 3.0 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.7 | 2.9 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.7 | 4.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.7 | 2.8 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.7 | 4.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.7 | 2.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.7 | 2.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.7 | 5.2 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.6 | 14.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.6 | 3.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 1.9 | GO:0097361 | CIA complex(GO:0097361) |
| 0.6 | 1.8 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.6 | 2.3 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.6 | 1.7 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.6 | 22.6 | GO:0005657 | replication fork(GO:0005657) |
| 0.5 | 1.6 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.5 | 4.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.5 | 5.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.5 | 4.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.4 | 3.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.4 | 4.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.4 | 121.3 | GO:0005764 | lysosome(GO:0005764) |
| 0.4 | 3.6 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.4 | 8.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.4 | 1.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.4 | 3.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.4 | 12.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.3 | 3.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 0.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.3 | 2.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.3 | 0.9 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 1.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.3 | 3.8 | GO:0005694 | chromosome(GO:0005694) |
| 0.3 | 0.9 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 1.5 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 1.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.3 | 1.9 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 1.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 3.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 7.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 1.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 1.8 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.2 | 6.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 7.4 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 3.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 4.9 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 5.3 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.2 | 1.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 1.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 8.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.2 | 5.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 27.6 | GO:0005819 | spindle(GO:0005819) |
| 0.2 | 2.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 1.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 2.0 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.2 | 2.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 5.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 1.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 2.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 12.2 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 2.0 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 1.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 6.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 2.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.2 | 3.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 3.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 2.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 7.0 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 1.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 2.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 4.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 7.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 2.9 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.1 | 5.0 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.0 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 4.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 13.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 1.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.5 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 4.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 4.0 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 9.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.4 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 1.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 26.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 0.3 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.6 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 3.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 20.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 6.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.3 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.4 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 3.6 | GO:0044427 | chromosomal part(GO:0044427) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 10.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 3.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 5.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 4.4 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 2.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 6.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 2.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 4.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 7.8 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 2.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 3.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 171.1 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 1.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.9 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 4.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.6 | GO:0044441 | ciliary part(GO:0044441) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 3.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 2.2 | 11.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 2.0 | 6.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.6 | 6.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 1.4 | 5.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.2 | 3.5 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 1.2 | 3.5 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 1.1 | 7.6 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.0 | 8.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 1.0 | 3.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 1.0 | 6.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.9 | 60.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.9 | 3.6 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.9 | 8.5 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.8 | 2.5 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.8 | 5.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.8 | 5.8 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.8 | 2.5 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.8 | 6.6 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.8 | 6.6 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.8 | 2.4 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.8 | 5.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.8 | 2.3 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.8 | 2.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.8 | 3.8 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.8 | 117.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.7 | 5.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.7 | 3.0 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.7 | 5.1 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.7 | 4.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.7 | 2.1 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.7 | 2.1 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.7 | 2.0 | GO:0032138 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.7 | 11.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.7 | 3.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.6 | 1.9 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.6 | 1.8 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.6 | 1.8 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.6 | 2.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.6 | 2.9 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.6 | 6.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.6 | 2.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.6 | 4.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.5 | 1.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.5 | 7.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.5 | 1.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.5 | 3.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 2.2 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.4 | 3.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.4 | 1.3 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.4 | 1.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 1.7 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.4 | 4.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 8.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 1.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.4 | 1.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.4 | 1.6 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.4 | 2.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.4 | 5.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.4 | 1.9 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.4 | 2.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.4 | 1.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.4 | 6.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 11.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.3 | 1.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 2.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 1.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 1.9 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 2.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.3 | 1.0 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 1.6 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 4.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.3 | 3.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 1.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.3 | 5.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 1.5 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.3 | 5.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.3 | 1.1 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.3 | 2.0 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.3 | 5.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.3 | 1.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 4.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 0.8 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.3 | 1.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 1.9 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.3 | 5.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 2.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.3 | 1.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 0.7 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.2 | 3.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.2 | 2.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.2 | 3.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 1.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 3.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 0.9 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.2 | 11.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 2.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 2.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 3.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 3.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 3.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 0.9 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.9 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 2.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 2.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.2 | 0.8 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.2 | 0.8 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 1.0 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 1.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 4.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 5.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 3.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 4.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 1.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 7.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 1.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 3.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 1.0 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 0.5 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.2 | 2.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 1.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 1.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 1.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 1.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 23.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.2 | 4.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 2.7 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 3.5 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.2 | 4.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 1.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.9 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 11.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 11.0 | GO:0051540 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.1 | 0.8 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 1.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 3.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 5.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.0 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 2.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 8.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 7.7 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 1.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0004532 | exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.1 | 4.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 1.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 2.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 6.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.0 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 1.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.5 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.1 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 0.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 5.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 23.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 10.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.9 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 1.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 10.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.6 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 1.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 2.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 1.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 4.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.9 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 1.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.8 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 2.8 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 1.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 3.0 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 0.6 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.9 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.8 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.3 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 1.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.1 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.3 | GO:0051916 | C-X-C chemokine receptor activity(GO:0016494) granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 6.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 11.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.6 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.3 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADPH binding(GO:0070402) NADH binding(GO:0070404) |
| 0.1 | 0.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 10.6 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 0.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.4 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.1 | 13.2 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 3.0 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 16.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 1.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 7.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 8.2 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 4.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 96.7 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 2.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.7 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.3 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 13.4 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.5 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 1.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.4 | GO:0030898 | microfilament motor activity(GO:0000146) actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 4.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 3.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.8 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0038187 | pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 70.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.9 | 29.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.8 | 28.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.6 | 5.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 8.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 4.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.4 | 9.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 18.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.3 | 3.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.3 | 18.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 9.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.3 | 5.0 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 3.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 1.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 1.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 9.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 3.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 2.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 3.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 3.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 4.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 3.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 2.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 5.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 6.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 9.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 4.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 3.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 2.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 8.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 5.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 25.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.2 | 24.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.0 | 7.9 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.8 | 6.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.8 | 6.4 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.8 | 8.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.7 | 10.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.7 | 16.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.7 | 18.6 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.6 | 2.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 6.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.5 | 13.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.5 | 22.8 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.5 | 8.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.5 | 3.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.5 | 3.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.4 | 2.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.4 | 25.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.4 | 3.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 10.9 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.3 | 4.4 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.3 | 13.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 1.6 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.3 | 4.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 3.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 4.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 1.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 2.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 5.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 4.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 3.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 3.0 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.2 | 4.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 3.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.2 | 2.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 0.9 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 8.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 2.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 1.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 1.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 11.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 3.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 7.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 6.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 2.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 4.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 3.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 0.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 14.9 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 0.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.6 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.1 | 1.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 3.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 0.8 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.0 | REACTOME INTERFERON SIGNALING | Genes involved in Interferon Signaling |
| 0.0 | 1.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.6 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |