Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
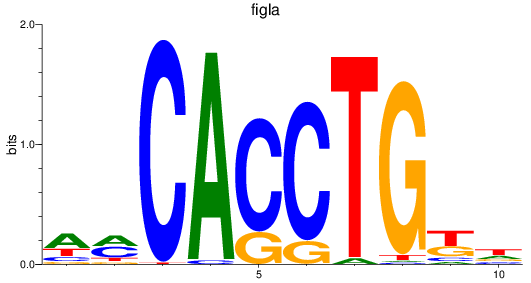
Results for figla
Z-value: 1.35
Transcription factors associated with figla
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
figla
|
ENSDARG00000087166 | folliculogenesis specific bHLH transcription factor |
|
figla
|
ENSDARG00000115572 | folliculogenesis specific bHLH transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| figla | dr11_v1_chr5_-_13848296_13848296 | 0.33 | 1.7e-01 | Click! |
Activity profile of figla motif
Sorted Z-values of figla motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_25472758 | 1.73 |
ENSDART00000011178
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr19_+_31904836 | 1.67 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr23_+_42414652 | 1.45 |
ENSDART00000171119
|
cyp2aa9
|
cytochrome P450, family 2, subfamily AA, polypeptide 9 |
| chr2_-_37893910 | 1.29 |
ENSDART00000143027
|
hbl2
|
hexose-binding lectin 2 |
| chr8_-_25247284 | 1.25 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr6_+_42338309 | 1.24 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr13_+_50375800 | 1.19 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr6_-_52348562 | 1.17 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr16_-_17207754 | 1.16 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr6_-_37459759 | 1.10 |
ENSDART00000190666
|
si:dkey-66a8.7
|
si:dkey-66a8.7 |
| chr18_-_14337065 | 1.09 |
ENSDART00000135703
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr19_-_30404096 | 1.02 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr9_+_159720 | 1.02 |
ENSDART00000166215
|
cldn10l2
|
claudin 10-like 2 |
| chr19_+_9174166 | 1.01 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr7_-_18881358 | 1.01 |
ENSDART00000021502
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr17_+_23462972 | 1.00 |
ENSDART00000112959
ENSDART00000192168 |
ankrd1a
|
ankyrin repeat domain 1a (cardiac muscle) |
| chr22_-_10470663 | 1.00 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr8_-_50287949 | 0.99 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr6_-_607063 | 0.99 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr6_-_37749711 | 0.99 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr8_-_8698607 | 0.95 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr4_+_5537101 | 0.95 |
ENSDART00000008692
|
si:dkey-14d8.7
|
si:dkey-14d8.7 |
| chr18_-_37241080 | 0.94 |
ENSDART00000126421
ENSDART00000078064 |
six9
|
SIX homeobox 9 |
| chr25_+_5012791 | 0.94 |
ENSDART00000156970
|
si:ch73-265h17.5
|
si:ch73-265h17.5 |
| chr12_-_28818720 | 0.91 |
ENSDART00000134453
ENSDART00000141727 |
prr15lb
|
proline rich 15-like b |
| chr4_-_8043839 | 0.90 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr8_-_39760427 | 0.89 |
ENSDART00000184336
ENSDART00000186500 |
si:ch211-170d8.8
|
si:ch211-170d8.8 |
| chr10_-_1961930 | 0.89 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr3_+_49021079 | 0.89 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr3_+_17910569 | 0.87 |
ENSDART00000080946
|
ttc25
|
tetratricopeptide repeat domain 25 |
| chr7_-_35066457 | 0.84 |
ENSDART00000058067
|
zgc:112160
|
zgc:112160 |
| chr9_+_29985010 | 0.81 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr18_+_21061216 | 0.80 |
ENSDART00000141739
|
fam169b
|
family with sequence similarity 169, member B |
| chr4_-_20483954 | 0.78 |
ENSDART00000187240
|
napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr10_-_1961576 | 0.78 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr19_-_30403922 | 0.77 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr16_+_17715243 | 0.76 |
ENSDART00000149437
ENSDART00000149596 |
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr18_+_25003207 | 0.75 |
ENSDART00000099476
ENSDART00000132285 |
fam174b
|
family with sequence similarity 174, member B |
| chr12_+_33484458 | 0.75 |
ENSDART00000000069
|
slc9a3r1a
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1a |
| chr20_+_18580176 | 0.74 |
ENSDART00000185310
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr5_+_28271412 | 0.74 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr20_+_38725662 | 0.74 |
ENSDART00000181873
|
uts1
|
urotensin 1 |
| chr21_-_17016788 | 0.72 |
ENSDART00000186703
|
AL844518.1
|
|
| chr10_+_33171501 | 0.72 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr15_+_36115955 | 0.72 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr15_+_19783352 | 0.71 |
ENSDART00000178784
ENSDART00000092560 |
zpld1b
|
zona pellucida-like domain containing 1b |
| chr23_+_42336084 | 0.71 |
ENSDART00000158959
ENSDART00000161812 |
cyp2aa7
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr21_+_11776328 | 0.71 |
ENSDART00000134469
ENSDART00000081646 |
glrx
|
glutaredoxin (thioltransferase) |
| chr6_-_37460178 | 0.71 |
ENSDART00000153500
|
si:dkey-66a8.7
|
si:dkey-66a8.7 |
| chr22_+_26400519 | 0.70 |
ENSDART00000159839
ENSDART00000144585 |
capn8
|
calpain 8 |
| chr14_-_16810401 | 0.70 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr20_+_36234335 | 0.70 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr2_+_37480669 | 0.69 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr23_+_25893020 | 0.69 |
ENSDART00000144769
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr3_+_34121156 | 0.69 |
ENSDART00000174929
|
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr14_+_32964 | 0.68 |
ENSDART00000166173
|
lyar
|
Ly1 antibody reactive homolog (mouse) |
| chr21_-_21178410 | 0.68 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr18_-_14337450 | 0.67 |
ENSDART00000061435
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr16_-_45854882 | 0.67 |
ENSDART00000027013
ENSDART00000128068 |
ntrk1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr22_+_19111444 | 0.67 |
ENSDART00000109655
|
fgf22
|
fibroblast growth factor 22 |
| chr11_+_31285127 | 0.67 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr8_-_30242706 | 0.66 |
ENSDART00000139864
ENSDART00000143809 |
zgc:162939
|
zgc:162939 |
| chr22_-_26524596 | 0.66 |
ENSDART00000087623
|
zgc:194330
|
zgc:194330 |
| chr6_-_54290227 | 0.66 |
ENSDART00000050483
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr22_-_17677947 | 0.66 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr13_-_12581388 | 0.66 |
ENSDART00000079655
|
enpep
|
glutamyl aminopeptidase |
| chr2_-_52795722 | 0.65 |
ENSDART00000193307
|
LO018490.1
|
|
| chr4_-_4834347 | 0.65 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr4_-_277081 | 0.64 |
ENSDART00000166174
|
si:ch73-252i11.1
|
si:ch73-252i11.1 |
| chr21_-_39931285 | 0.64 |
ENSDART00000180010
ENSDART00000024407 |
tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr19_-_35035857 | 0.63 |
ENSDART00000103253
|
bmp8a
|
bone morphogenetic protein 8a |
| chr4_-_8040436 | 0.63 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr9_+_24121036 | 0.62 |
ENSDART00000101565
|
maip1
|
matrix AAA peptidase interacting protein 1 |
| chr2_-_37888429 | 0.61 |
ENSDART00000183277
|
mbl2
|
mannose binding lectin 2 |
| chr8_+_27555314 | 0.61 |
ENSDART00000135568
ENSDART00000016696 |
rhocb
|
ras homolog family member Cb |
| chr6_+_42918933 | 0.61 |
ENSDART00000064896
|
gnat1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
| chr17_+_5976683 | 0.61 |
ENSDART00000110276
|
zgc:194275
|
zgc:194275 |
| chr15_-_29326254 | 0.61 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr6_+_40775800 | 0.59 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr13_-_46991577 | 0.58 |
ENSDART00000114748
|
vip
|
vasoactive intestinal peptide |
| chr8_+_13849999 | 0.58 |
ENSDART00000143784
|
doc2d
|
double C2-like domains, delta |
| chr7_-_5640822 | 0.58 |
ENSDART00000143767
|
BX000524.1
|
|
| chr11_+_34921492 | 0.58 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr5_-_15494164 | 0.57 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr7_-_50367326 | 0.57 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr14_+_22297257 | 0.56 |
ENSDART00000113676
|
atp10b
|
ATPase phospholipid transporting 10B |
| chr13_+_40770628 | 0.56 |
ENSDART00000085846
|
nkx1.2la
|
NK1 transcription factor related 2-like,a |
| chr21_+_33172526 | 0.56 |
ENSDART00000183532
|
arl3l1
|
ADP-ribosylation factor-like 3, like 1 |
| chr3_+_14641962 | 0.56 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr21_+_34981263 | 0.56 |
ENSDART00000132711
|
rbm11
|
RNA binding motif protein 11 |
| chr11_+_7264457 | 0.56 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr12_-_26415499 | 0.56 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr19_-_31341850 | 0.56 |
ENSDART00000040810
|
arl4ab
|
ADP-ribosylation factor-like 4ab |
| chr7_+_58751504 | 0.55 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr13_+_24834199 | 0.55 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr12_-_13336703 | 0.55 |
ENSDART00000134356
|
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr6_-_55423220 | 0.54 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr7_+_33279618 | 0.54 |
ENSDART00000173706
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr21_-_25774605 | 0.53 |
ENSDART00000002128
|
CLDN4 (1 of many)
|
zgc:112437 |
| chr4_-_16001118 | 0.52 |
ENSDART00000041070
ENSDART00000125389 |
mest
|
mesoderm specific transcript |
| chr1_-_1894722 | 0.52 |
ENSDART00000165669
|
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr19_-_10612774 | 0.51 |
ENSDART00000091898
|
si:dkey-211g8.6
|
si:dkey-211g8.6 |
| chr12_-_4301234 | 0.51 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr22_-_5006801 | 0.50 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr14_-_7409364 | 0.50 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr14_+_22172047 | 0.50 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr25_-_24240797 | 0.49 |
ENSDART00000132790
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr7_+_57089354 | 0.49 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr2_-_50225411 | 0.47 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr14_+_34514336 | 0.47 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr6_-_33129045 | 0.47 |
ENSDART00000073757
|
tspan1
|
tetraspanin 1 |
| chr24_+_8736497 | 0.47 |
ENSDART00000181904
|
tmem14ca
|
transmembrane protein 14Ca |
| chr21_+_30351256 | 0.47 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr25_+_27744293 | 0.47 |
ENSDART00000103519
ENSDART00000149456 |
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr8_-_13029297 | 0.46 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr5_-_69212184 | 0.46 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr23_-_1583193 | 0.46 |
ENSDART00000143841
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr17_-_15777350 | 0.46 |
ENSDART00000155711
|
ankrd6a
|
ankyrin repeat domain 6a |
| chr20_+_21391181 | 0.46 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr5_-_29152457 | 0.45 |
ENSDART00000078469
|
noxa1
|
NADPH oxidase activator 1 |
| chr15_+_33989181 | 0.45 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr20_+_36623807 | 0.45 |
ENSDART00000149171
ENSDART00000062895 |
srp9
|
signal recognition particle 9 |
| chr2_+_34967022 | 0.44 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr8_-_14554785 | 0.44 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr6_+_49028874 | 0.44 |
ENSDART00000175254
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr1_-_53468160 | 0.44 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr8_-_11229523 | 0.44 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr2_+_34967210 | 0.43 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr21_+_27336550 | 0.43 |
ENSDART00000139634
|
dpp3
|
dipeptidyl-peptidase 3 |
| chr16_+_54875530 | 0.43 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr1_+_594584 | 0.42 |
ENSDART00000135944
|
jam2a
|
junctional adhesion molecule 2a |
| chr25_-_9013963 | 0.42 |
ENSDART00000073912
|
rag2
|
recombination activating gene 2 |
| chr5_-_25572151 | 0.42 |
ENSDART00000144995
|
si:dkey-229d2.4
|
si:dkey-229d2.4 |
| chr6_-_8360918 | 0.42 |
ENSDART00000004716
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr14_+_2487672 | 0.42 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr20_-_43743700 | 0.41 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr22_+_7439186 | 0.41 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr9_+_50749388 | 0.41 |
ENSDART00000181015
|
fign
|
fidgetin |
| chr7_+_69841017 | 0.41 |
ENSDART00000169107
|
FO818704.1
|
|
| chr21_-_25522510 | 0.41 |
ENSDART00000162711
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr14_-_33277743 | 0.41 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr15_+_38299385 | 0.40 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr19_-_9711472 | 0.40 |
ENSDART00000016197
ENSDART00000175075 |
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr3_+_36515376 | 0.40 |
ENSDART00000161652
|
si:dkeyp-72e1.9
|
si:dkeyp-72e1.9 |
| chr4_+_5255041 | 0.40 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr15_+_24572926 | 0.40 |
ENSDART00000155636
ENSDART00000187800 |
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr23_-_19831739 | 0.40 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr16_-_42066523 | 0.40 |
ENSDART00000180538
ENSDART00000058620 |
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr12_+_30705769 | 0.39 |
ENSDART00000186448
ENSDART00000066259 |
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr5_-_22052852 | 0.39 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr24_+_35564668 | 0.39 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr23_+_4741543 | 0.39 |
ENSDART00000144761
|
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a |
| chr8_+_44358443 | 0.39 |
ENSDART00000189130
ENSDART00000189212 |
CU914776.2
|
|
| chr20_+_18703454 | 0.39 |
ENSDART00000152342
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr7_-_20836625 | 0.39 |
ENSDART00000192566
|
cldn15a
|
claudin 15a |
| chr20_-_39188476 | 0.38 |
ENSDART00000152858
|
rcan2
|
regulator of calcineurin 2 |
| chr10_-_38468847 | 0.38 |
ENSDART00000133914
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr6_+_50452699 | 0.38 |
ENSDART00000115049
|
mych
|
myelocytomatosis oncogene homolog |
| chr19_-_27542433 | 0.38 |
ENSDART00000136414
|
si:ch211-152p11.4
|
si:ch211-152p11.4 |
| chr25_-_34632050 | 0.38 |
ENSDART00000170671
|
megf8
|
multiple EGF-like-domains 8 |
| chr4_-_12978925 | 0.38 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr18_+_8402129 | 0.37 |
ENSDART00000081154
ENSDART00000171974 |
prpf18
|
PRP18 pre-mRNA processing factor 18 homolog (yeast) |
| chr12_+_30706158 | 0.37 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr10_-_31805923 | 0.36 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr23_-_26532696 | 0.36 |
ENSDART00000124811
ENSDART00000180274 |
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr13_+_24584401 | 0.36 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr15_-_23761580 | 0.36 |
ENSDART00000137918
|
bbc3
|
BCL2 binding component 3 |
| chr14_-_33278084 | 0.35 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr8_-_22273819 | 0.35 |
ENSDART00000121513
|
nphp4
|
nephronophthisis 4 |
| chr17_-_8692722 | 0.35 |
ENSDART00000148931
ENSDART00000192891 |
ctbp2a
|
C-terminal binding protein 2a |
| chr11_-_5865744 | 0.34 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr6_-_41135215 | 0.34 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr11_+_31730680 | 0.34 |
ENSDART00000145497
|
diaph3
|
diaphanous-related formin 3 |
| chr5_-_37900350 | 0.34 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr3_-_32169754 | 0.34 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr8_+_14158021 | 0.34 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr23_+_30985530 | 0.33 |
ENSDART00000142953
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr5_+_58679071 | 0.33 |
ENSDART00000019561
|
zgc:171734
|
zgc:171734 |
| chr23_-_15090782 | 0.33 |
ENSDART00000133624
|
si:ch211-218g4.2
|
si:ch211-218g4.2 |
| chr1_-_59141715 | 0.33 |
ENSDART00000164941
ENSDART00000138870 |
si:ch1073-110a20.1
|
si:ch1073-110a20.1 |
| chr18_+_34861568 | 0.33 |
ENSDART00000192825
|
LO018333.1
|
|
| chr16_+_32152612 | 0.33 |
ENSDART00000008880
|
gprc6a
|
G protein-coupled receptor, class C, group 6, member A |
| chr20_+_46371458 | 0.33 |
ENSDART00000152912
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr7_+_4524059 | 0.33 |
ENSDART00000187693
|
si:dkey-83f18.4
|
si:dkey-83f18.4 |
| chr6_+_11438972 | 0.32 |
ENSDART00000029314
|
col5a2b
|
collagen, type V, alpha 2b |
| chr6_-_12644563 | 0.32 |
ENSDART00000153797
|
dock9b
|
dedicator of cytokinesis 9b |
| chr11_-_30352333 | 0.32 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr23_+_9522942 | 0.32 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr5_+_40485503 | 0.32 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr4_+_10066840 | 0.31 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr18_+_7345417 | 0.31 |
ENSDART00000041429
|
glipr1b
|
GLI pathogenesis-related 1b |
| chr17_-_25331439 | 0.31 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr19_-_34927201 | 0.31 |
ENSDART00000076518
|
sla1
|
Src-like-adaptor 1 |
| chr10_-_25217347 | 0.31 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr22_-_18546649 | 0.31 |
ENSDART00000171277
|
cirbpb
|
cold inducible RNA binding protein b |
| chr15_-_34878388 | 0.31 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr23_+_21479958 | 0.31 |
ENSDART00000188302
ENSDART00000144320 |
si:dkey-1c11.1
|
si:dkey-1c11.1 |
| chr13_-_438705 | 0.30 |
ENSDART00000082142
|
CU570800.1
|
|
| chr6_-_33129312 | 0.30 |
ENSDART00000156987
|
tspan1
|
tetraspanin 1 |
| chr2_-_1227221 | 0.30 |
ENSDART00000130897
|
ABCF3
|
ATP binding cassette subfamily F member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of figla
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.4 | 1.2 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 0.9 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.2 | 1.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.2 | 0.8 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.2 | 1.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 0.5 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.6 | GO:0033605 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.1 | 0.7 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.2 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.1 | 0.7 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.6 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.4 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.1 | 1.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.2 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.8 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 2.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.3 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 | 0.7 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.6 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.3 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.5 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.7 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.4 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.2 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 0.4 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 1.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.7 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.5 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.2 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 1.0 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 0.7 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 2.4 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.5 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.4 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.0 | 1.0 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.4 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.6 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 1.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 1.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.1 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly involved in wound healing(GO:0090505) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.4 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.7 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.5 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 0.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.1 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.5 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.4 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.6 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 0.5 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.2 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.4 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 2.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 2.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.6 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.9 | GO:0030141 | secretory granule(GO:0030141) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 0.6 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 1.7 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.5 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.5 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 2.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.4 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 1.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 2.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 1.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 2.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.7 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 0.7 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 1.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 0.6 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 2.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |