Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
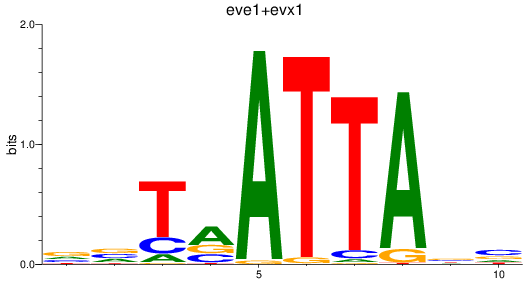
Results for eve1+evx1
Z-value: 0.86
Transcription factors associated with eve1+evx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
eve1
|
ENSDARG00000056012 | even-skipped-like1 |
|
evx1
|
ENSDARG00000099365 | even-skipped homeobox 1 |
|
evx1
|
ENSDARG00000100087 | even-skipped homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| evx1 | dr11_v1_chr19_-_19871211_19871211 | -0.78 | 7.8e-05 | Click! |
| eve1 | dr11_v1_chr3_-_23643751_23643830 | -0.49 | 3.4e-02 | Click! |
Activity profile of eve1+evx1 motif
Sorted Z-values of eve1+evx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_11385155 | 4.26 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr19_+_40856534 | 3.83 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr18_+_1837668 | 3.68 |
ENSDART00000164210
|
CABZ01079192.1
|
|
| chr19_+_40856807 | 3.38 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr3_-_31079186 | 2.84 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr8_-_21372446 | 2.46 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr4_+_11384891 | 2.35 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr16_-_54455573 | 2.22 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr23_+_40460333 | 2.05 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr8_+_53423408 | 2.02 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr21_-_27185915 | 1.97 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr6_-_55585423 | 1.91 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr9_-_6372535 | 1.85 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr25_-_25384045 | 1.75 |
ENSDART00000150631
|
zgc:123278
|
zgc:123278 |
| chr18_+_9615147 | 1.71 |
ENSDART00000160284
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr23_-_19230627 | 1.69 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr20_-_40755614 | 1.65 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr10_+_25726694 | 1.58 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr2_-_1443692 | 1.56 |
ENSDART00000004533
|
rpe65a
|
retinal pigment epithelium-specific protein 65a |
| chr11_+_38280454 | 1.52 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr1_-_47071979 | 1.48 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr19_-_657439 | 1.42 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr4_-_67969695 | 1.40 |
ENSDART00000190016
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr1_+_17676745 | 1.36 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr5_+_64732270 | 1.33 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr10_+_3049636 | 1.27 |
ENSDART00000081794
ENSDART00000183167 ENSDART00000191634 ENSDART00000183514 |
rasgrf2a
|
Ras protein-specific guanine nucleotide-releasing factor 2a |
| chr25_+_3327071 | 1.26 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr18_+_11506561 | 1.24 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr5_+_64732036 | 1.21 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr24_-_39518599 | 1.20 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr15_+_9053059 | 1.17 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr15_+_47418565 | 1.17 |
ENSDART00000155709
|
clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr21_+_30355767 | 1.17 |
ENSDART00000189948
|
CR749164.1
|
|
| chr3_+_1005059 | 1.16 |
ENSDART00000132460
|
zgc:172051
|
zgc:172051 |
| chr13_+_38814521 | 1.10 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr17_-_23616626 | 1.09 |
ENSDART00000104730
|
ifit14
|
interferon-induced protein with tetratricopeptide repeats 14 |
| chr21_+_1381276 | 1.07 |
ENSDART00000192907
|
TCF4
|
transcription factor 4 |
| chr2_-_57076687 | 1.04 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr23_+_33963619 | 1.03 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr23_-_44219902 | 1.03 |
ENSDART00000185874
|
zgc:158659
|
zgc:158659 |
| chr22_+_17205608 | 1.00 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr1_+_11977426 | 0.99 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr2_+_31492662 | 0.99 |
ENSDART00000123495
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr17_-_28198099 | 0.96 |
ENSDART00000156143
|
htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr21_-_3770636 | 0.95 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr16_-_2414063 | 0.94 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr20_-_46362606 | 0.92 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr18_-_43884044 | 0.90 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr5_+_27137473 | 0.84 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr19_-_205104 | 0.84 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr2_-_32825917 | 0.84 |
ENSDART00000180409
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr15_-_34785594 | 0.84 |
ENSDART00000154256
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr10_+_41765944 | 0.84 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr9_+_17983463 | 0.84 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr2_-_16380283 | 0.83 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr24_-_1341543 | 0.83 |
ENSDART00000169341
|
nrp1a
|
neuropilin 1a |
| chr19_+_5543072 | 0.83 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr5_-_30615901 | 0.80 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr23_+_26946744 | 0.79 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr23_+_4646194 | 0.79 |
ENSDART00000092344
|
LO017700.1
|
|
| chr21_+_11244068 | 0.78 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr11_+_45323455 | 0.77 |
ENSDART00000189249
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr7_-_24046999 | 0.77 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr16_+_5774977 | 0.77 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr10_+_21867307 | 0.75 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr21_-_18993110 | 0.74 |
ENSDART00000144086
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr6_+_24398907 | 0.73 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr19_+_712127 | 0.73 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr16_+_17389116 | 0.73 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr21_-_3853204 | 0.72 |
ENSDART00000188829
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr2_-_32826108 | 0.72 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr2_-_11258547 | 0.69 |
ENSDART00000165803
ENSDART00000193817 |
slc44a5a
|
solute carrier family 44, member 5a |
| chr4_+_12612723 | 0.69 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr12_-_32426685 | 0.69 |
ENSDART00000105574
|
enpp7.2
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 2 |
| chr14_-_8625845 | 0.67 |
ENSDART00000054760
|
zgc:162144
|
zgc:162144 |
| chr6_+_9870192 | 0.67 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr3_-_34084387 | 0.66 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr18_+_28106139 | 0.66 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr8_+_52503340 | 0.66 |
ENSDART00000168838
|
si:ch1073-392o20.1
|
si:ch1073-392o20.1 |
| chr11_+_28476298 | 0.65 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr6_+_41039166 | 0.65 |
ENSDART00000125659
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr9_-_21918963 | 0.64 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr11_+_36231248 | 0.63 |
ENSDART00000131104
|
GPR62 (1 of many)
|
si:ch211-213o11.11 |
| chr1_-_10647484 | 0.63 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr20_+_22067337 | 0.62 |
ENSDART00000152636
|
clocka
|
clock circadian regulator a |
| chr4_-_49582108 | 0.62 |
ENSDART00000154999
|
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr1_+_16396870 | 0.61 |
ENSDART00000189245
ENSDART00000113266 |
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr23_-_33361425 | 0.59 |
ENSDART00000031638
|
slc48a1a
|
solute carrier family 48 (heme transporter), member 1a |
| chr6_-_6487876 | 0.56 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr19_+_9033376 | 0.55 |
ENSDART00000192298
ENSDART00000052915 |
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr8_-_30338872 | 0.53 |
ENSDART00000137583
|
dock8
|
dedicator of cytokinesis 8 |
| chr24_+_36636208 | 0.53 |
ENSDART00000139211
|
si:ch73-334d15.4
|
si:ch73-334d15.4 |
| chr5_+_19314574 | 0.51 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr1_-_9195629 | 0.51 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr5_-_42904329 | 0.47 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr1_-_46981134 | 0.46 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr21_-_5205617 | 0.46 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr12_-_24060358 | 0.46 |
ENSDART00000050831
|
chac2
|
ChaC, cation transport regulator homolog 2 (E. coli) |
| chr18_+_50461981 | 0.45 |
ENSDART00000158761
|
CU896640.1
|
|
| chr2_+_21229909 | 0.44 |
ENSDART00000046127
|
vipr1a
|
vasoactive intestinal peptide receptor 1a |
| chr15_-_22074315 | 0.44 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr8_-_21142550 | 0.43 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr19_-_44054930 | 0.43 |
ENSDART00000151084
ENSDART00000150991 ENSDART00000005191 |
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chrM_+_12897 | 0.43 |
ENSDART00000093622
|
mt-nd5
|
NADH dehydrogenase 5, mitochondrial |
| chr1_+_51191049 | 0.41 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr5_+_38263240 | 0.41 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr16_-_36093776 | 0.41 |
ENSDART00000159134
|
OSCP1
|
organic solute carrier partner 1 |
| chr16_-_25829779 | 0.40 |
ENSDART00000086301
|
irge4
|
immunity-related GTPase family, e4 |
| chr3_+_54047342 | 0.40 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr6_+_515181 | 0.40 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr18_-_1185772 | 0.40 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr18_+_11970987 | 0.39 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr10_-_21955231 | 0.39 |
ENSDART00000183695
|
FO744833.2
|
|
| chr3_-_8399774 | 0.38 |
ENSDART00000190238
ENSDART00000113140 ENSDART00000132949 |
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr24_-_41320037 | 0.37 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr6_+_39232245 | 0.36 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr7_-_64589920 | 0.35 |
ENSDART00000172619
ENSDART00000184113 |
CR387919.1
|
|
| chr21_+_40292801 | 0.34 |
ENSDART00000174220
|
si:ch211-218m3.16
|
si:ch211-218m3.16 |
| chr23_-_20051369 | 0.33 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr7_+_30787903 | 0.32 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr14_+_45406299 | 0.31 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr3_+_32832538 | 0.31 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr3_+_17537352 | 0.30 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr22_-_37686966 | 0.30 |
ENSDART00000192217
|
htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr19_+_19241372 | 0.30 |
ENSDART00000184392
ENSDART00000165008 |
ptpn23b
|
protein tyrosine phosphatase, non-receptor type 23, b |
| chr15_+_22014029 | 0.30 |
ENSDART00000079504
|
ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr23_-_24394719 | 0.30 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr1_-_10647307 | 0.30 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr7_-_27685365 | 0.30 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr4_+_9400012 | 0.29 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr3_+_27786601 | 0.28 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr12_-_19007834 | 0.27 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr11_-_44876005 | 0.26 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr8_-_27656765 | 0.26 |
ENSDART00000078491
|
mov10b.2
|
Moloney leukemia virus 10b, tandem duplicate 2 |
| chr15_-_23793641 | 0.26 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr18_-_12052132 | 0.26 |
ENSDART00000074361
|
zgc:110789
|
zgc:110789 |
| chr20_-_9462433 | 0.25 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr25_+_26923193 | 0.25 |
ENSDART00000187364
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr12_-_4408828 | 0.24 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr20_-_13722514 | 0.24 |
ENSDART00000152458
|
si:ch211-122h15.4
|
si:ch211-122h15.4 |
| chr2_-_21819421 | 0.23 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr1_+_16397063 | 0.23 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr1_-_17715493 | 0.23 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr22_-_10121880 | 0.21 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr25_+_35891342 | 0.20 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr2_-_17235891 | 0.19 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr11_-_45152702 | 0.19 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr9_+_41156818 | 0.19 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr3_+_18398876 | 0.18 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr16_-_21038015 | 0.18 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr6_-_57655030 | 0.18 |
ENSDART00000155438
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr8_-_24252933 | 0.17 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr14_+_924876 | 0.17 |
ENSDART00000183908
|
myoz3a
|
myozenin 3a |
| chr2_+_30464829 | 0.15 |
ENSDART00000181432
|
neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr5_+_26765275 | 0.14 |
ENSDART00000144169
|
si:ch211-102c2.8
|
si:ch211-102c2.8 |
| chr21_+_34814444 | 0.14 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr10_-_41664427 | 0.12 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr4_-_71912457 | 0.12 |
ENSDART00000179685
|
si:dkey-92c21.1
|
si:dkey-92c21.1 |
| chr2_+_30465102 | 0.12 |
ENSDART00000188404
|
neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr16_+_11029762 | 0.10 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr21_-_42831033 | 0.09 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr22_-_881725 | 0.08 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr20_+_41021054 | 0.08 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr11_-_29768054 | 0.08 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr2_-_26720854 | 0.08 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr15_+_29662401 | 0.07 |
ENSDART00000135540
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr22_+_20427170 | 0.07 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr17_-_23446760 | 0.06 |
ENSDART00000104715
|
pcgf5a
|
polycomb group ring finger 5a |
| chr9_-_54361357 | 0.06 |
ENSDART00000149813
|
rad50
|
RAD50 homolog, double strand break repair protein |
| chr13_+_33117528 | 0.06 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr1_+_55758257 | 0.06 |
ENSDART00000139312
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr15_+_31481939 | 0.05 |
ENSDART00000134306
|
or102-5
|
odorant receptor, family C, subfamily 102, member 5 |
| chr15_+_5360407 | 0.05 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr16_-_29458806 | 0.05 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr3_+_13624815 | 0.03 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr6_-_40352215 | 0.02 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr2_-_15324837 | 0.02 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr24_+_16547035 | 0.00 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
Network of associatons between targets according to the STRING database.
First level regulatory network of eve1+evx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.4 | 4.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.4 | 1.6 | GO:0016123 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.4 | 1.1 | GO:0044038 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.3 | 1.4 | GO:0051503 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.3 | 2.5 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.2 | 0.8 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.2 | 0.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 2.0 | GO:0042044 | fluid transport(GO:0042044) |
| 0.1 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.8 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.4 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 1.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.5 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 1.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.3 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 2.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.6 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 1.3 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.1 | 2.8 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 0.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.3 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 1.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 1.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.9 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 2.2 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.2 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 1.0 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 2.0 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.7 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.6 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.1 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.0 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.4 | 7.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.4 | 4.1 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 0.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.6 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.8 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 1.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 3.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 3.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.4 | 2.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.0 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 7.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 1.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.2 | 4.1 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 1.7 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 1.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 2.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.5 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 3.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.3 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.3 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.9 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.2 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.1 | 0.4 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.0 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.7 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.4 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 1.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 1.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 1.0 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 1.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |