Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
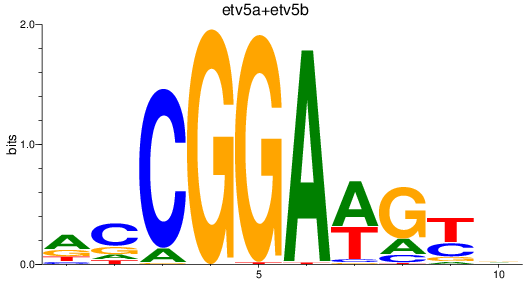
Results for etv5a+etv5b_elk1_elk4_etv1_elf1_erf+erfl3_gabpa_elk3
Z-value: 6.17
Transcription factors associated with etv5a+etv5b_elk1_elk4_etv1_elf1_erf+erfl3_gabpa_elk3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
etv5b
|
ENSDARG00000044511 | ETS variant transcription factor 5b |
|
etv5a
|
ENSDARG00000069763 | ETS variant transcription factor 5a |
|
etv5a
|
ENSDARG00000113729 | ETS variant transcription factor 5a |
|
etv5b
|
ENSDARG00000113744 | ETS variant transcription factor 5b |
|
elk1
|
ENSDARG00000078066 | ETS transcription factor ELK1 |
|
elk4
|
ENSDARG00000077092 | ETS transcription factor ELK4 |
|
etv1
|
ENSDARG00000101959 | ETS variant transcription factor 1 |
|
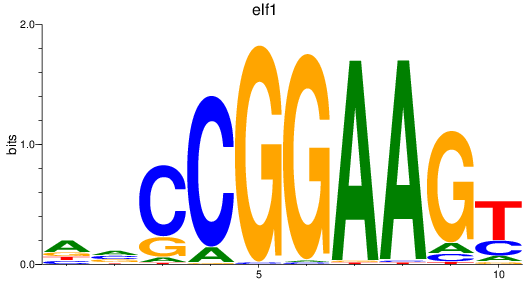
elf1
|
ENSDARG00000020759 | E74-like ETS transcription factor 1 |
|
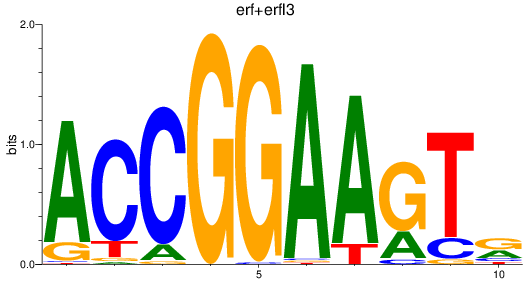
erfl3
|
ENSDARG00000062801 | Ets2 repressor factor like 3 |
|
erf
|
ENSDARG00000063417 | Ets2 repressor factor |
|
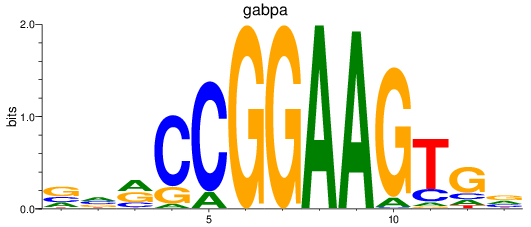
gabpa
|
ENSDARG00000069289 | GA binding protein transcription factor subunit alpha |
|
gabpa
|
ENSDARG00000110923 | GA binding protein transcription factor subunit alpha |
|
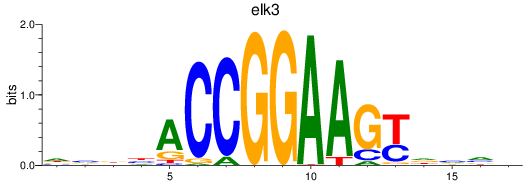
elk3
|
ENSDARG00000018688 | ETS transcription factor ELK3 |
|
elk3
|
ENSDARG00000110853 | ETS transcription factor ELK3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| etv1 | dr11_v1_chr15_-_34214440_34214440 | -0.94 | 1.8e-09 | Click! |
| gabpa | dr11_v1_chr9_+_35016201_35016201 | 0.93 | 5.2e-09 | Click! |
| elk1 | dr11_v1_chr8_+_8735285_8735285 | -0.92 | 2.3e-08 | Click! |
| etv5a | dr11_v1_chr9_+_22632126_22632126 | -0.80 | 4.0e-05 | Click! |
| elk3 | dr11_v1_chr4_+_7677318_7677318 | 0.78 | 8.4e-05 | Click! |
| elk4 | dr11_v1_chr11_-_38492564_38492564 | -0.73 | 4.3e-04 | Click! |
| etv5b | dr11_v1_chr6_-_14040136_14040161 | 0.69 | 1.0e-03 | Click! |
| erf | dr11_v1_chr19_-_6193067_6193141 | 0.68 | 1.3e-03 | Click! |
| elf1 | dr11_v1_chr14_-_40821411_40821411 | -0.47 | 4.4e-02 | Click! |
| erfl3 | dr11_v1_chr16_+_11029762_11029762 | -0.44 | 6.2e-02 | Click! |
Activity profile of etv5a+etv5b_elk1_elk4_etv1_elf1_erf+erfl3_gabpa_elk3 motif
Sorted Z-values of etv5a+etv5b_elk1_elk4_etv1_elf1_erf+erfl3_gabpa_elk3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_36554675 | 25.02 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr4_+_12031958 | 21.26 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr8_-_41228530 | 19.42 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr13_-_33700461 | 15.28 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr8_+_48943009 | 14.93 |
ENSDART00000180763
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr25_+_186583 | 14.86 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr5_+_28497956 | 14.76 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr1_-_21723329 | 14.55 |
ENSDART00000137138
|
si:ch211-134c9.2
|
si:ch211-134c9.2 |
| chr20_+_13141408 | 14.42 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr13_+_15933168 | 14.33 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr25_-_19574146 | 14.04 |
ENSDART00000156811
|
si:ch211-59o9.10
|
si:ch211-59o9.10 |
| chr12_-_49151326 | 13.55 |
ENSDART00000153244
|
bub3
|
BUB3 mitotic checkpoint protein |
| chr14_-_14687004 | 13.38 |
ENSDART00000169970
|
gcna
|
germ cell nuclear acidic peptidase |
| chr11_-_16115804 | 13.27 |
ENSDART00000143436
ENSDART00000157928 |
rpf1
|
ribosome production factor 1 homolog |
| chr12_+_10443785 | 13.22 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr16_+_20294976 | 13.16 |
ENSDART00000059619
|
fkbp14
|
FK506 binding protein 14 |
| chr18_+_29898955 | 13.16 |
ENSDART00000064080
|
cenpn
|
centromere protein N |
| chr7_+_756942 | 12.56 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr5_-_56119028 | 12.54 |
ENSDART00000083134
|
mks1
|
Meckel syndrome, type 1 |
| chr9_+_426392 | 12.53 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr7_+_55518519 | 12.49 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr14_-_31618243 | 12.44 |
ENSDART00000016592
|
mmgt1
|
membrane magnesium transporter 1 |
| chr5_+_69733096 | 12.42 |
ENSDART00000169013
|
arl6ip4
|
ADP-ribosylation factor-like 6 interacting protein 4 |
| chr3_-_14498295 | 12.39 |
ENSDART00000172102
|
elof1
|
ELF1 homolog, elongation factor 1 |
| chr5_+_3927989 | 12.32 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr5_-_67365006 | 12.26 |
ENSDART00000136116
|
unga
|
uracil DNA glycosylase a |
| chr22_-_28373698 | 11.83 |
ENSDART00000157592
|
si:ch211-213c4.5
|
si:ch211-213c4.5 |
| chr13_-_29980215 | 11.55 |
ENSDART00000042049
|
hif1an
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr7_-_57637779 | 11.40 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr5_-_51998708 | 11.25 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr4_+_58576146 | 11.22 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr20_-_33497128 | 11.21 |
ENSDART00000101974
|
erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr8_+_36554816 | 11.20 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr23_-_13840433 | 11.19 |
ENSDART00000104831
|
naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr21_-_30408775 | 11.10 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr4_+_9592486 | 11.08 |
ENSDART00000080829
|
hspa14
|
heat shock protein 14 |
| chr21_-_45840341 | 11.03 |
ENSDART00000006942
|
irf1b
|
interferon regulatory factor 1b |
| chr14_-_10617923 | 10.88 |
ENSDART00000133723
ENSDART00000131939 ENSDART00000136649 |
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr18_-_26797723 | 10.79 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr13_+_41917606 | 10.71 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr22_-_10752471 | 10.63 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr16_+_52966812 | 10.57 |
ENSDART00000148435
ENSDART00000049099 ENSDART00000150117 |
trip13
|
thyroid hormone receptor interactor 13 |
| chr2_-_8688759 | 10.54 |
ENSDART00000010257
|
miga1
|
mitoguardin 1 |
| chr24_-_36175365 | 10.51 |
ENSDART00000065338
|
pak1ip1
|
PAK1 interacting protein 1 |
| chr18_-_3552414 | 10.48 |
ENSDART00000163762
ENSDART00000165434 ENSDART00000161197 ENSDART00000166841 ENSDART00000170260 |
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr7_+_32693890 | 10.44 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr20_-_14462995 | 10.38 |
ENSDART00000152418
ENSDART00000044125 |
grcc10
|
gene rich cluster, C10 gene |
| chr2_+_38055529 | 10.27 |
ENSDART00000145642
|
si:rp71-1g18.1
|
si:rp71-1g18.1 |
| chr3_+_59880317 | 10.19 |
ENSDART00000166922
ENSDART00000108647 |
alyref
|
Aly/REF export factor |
| chr18_-_7056702 | 10.13 |
ENSDART00000127397
ENSDART00000148774 |
utp15
|
utp15, U3 small nucleolar ribonucleoprotein, homolog |
| chr8_-_17184482 | 10.10 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr12_-_34435604 | 10.05 |
ENSDART00000115088
|
birc5a
|
baculoviral IAP repeat containing 5a |
| chr19_+_3206263 | 9.97 |
ENSDART00000020344
|
zgc:86598
|
zgc:86598 |
| chr16_-_54498109 | 9.96 |
ENSDART00000083713
|
clk2b
|
CDC-like kinase 2b |
| chr2_+_30182431 | 9.95 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr21_-_226071 | 9.94 |
ENSDART00000160667
|
nup54
|
nucleoporin 54 |
| chr25_+_3549401 | 9.90 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr14_+_4276394 | 9.88 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr11_-_36230146 | 9.78 |
ENSDART00000135888
ENSDART00000189782 |
rrp9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr4_+_9536860 | 9.72 |
ENSDART00000130083
|
lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr12_-_28363111 | 9.70 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr15_-_14064302 | 9.69 |
ENSDART00000156384
|
BX927244.1
|
|
| chr19_-_34089205 | 9.67 |
ENSDART00000163618
ENSDART00000161666 |
rp9
|
RP9, pre-mRNA splicing factor |
| chr5_-_67365750 | 9.63 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr19_-_37508571 | 9.62 |
ENSDART00000018255
|
ilf2
|
interleukin enhancer binding factor 2 |
| chr20_-_2619316 | 9.60 |
ENSDART00000185777
|
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr5_-_67365333 | 9.59 |
ENSDART00000133438
|
unga
|
uracil DNA glycosylase a |
| chr4_-_330748 | 9.53 |
ENSDART00000067488
|
mrps10
|
mitochondrial ribosomal protein S10 |
| chr15_-_43625549 | 9.47 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr22_+_10752787 | 9.35 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr17_+_43013171 | 9.20 |
ENSDART00000055541
|
gskip
|
gsk3b interacting protein |
| chr2_+_8649293 | 9.13 |
ENSDART00000081442
ENSDART00000186024 |
fubp1
|
far upstream element (FUSE) binding protein 1 |
| chr22_+_10752511 | 9.12 |
ENSDART00000081188
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_-_3927692 | 9.11 |
ENSDART00000146840
ENSDART00000058346 |
c1qbp
|
complement component 1, q subcomponent binding protein |
| chr23_-_31403668 | 9.10 |
ENSDART00000147498
|
CR759830.1
|
|
| chr9_-_30555725 | 9.03 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr21_-_26715270 | 9.02 |
ENSDART00000053794
|
banf1
|
barrier to autointegration factor 1 |
| chr21_+_17016337 | 8.99 |
ENSDART00000065755
|
gpn3
|
GPN-loop GTPase 3 |
| chr9_+_48819280 | 8.99 |
ENSDART00000112555
|
spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr19_+_34742706 | 8.95 |
ENSDART00000103276
|
fam206a
|
family with sequence similarity 206, member A |
| chr20_-_42972599 | 8.94 |
ENSDART00000100751
|
pomcb
|
proopiomelanocortin b |
| chr13_+_33655404 | 8.84 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr2_-_32512648 | 8.79 |
ENSDART00000170674
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr19_-_18855513 | 8.77 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr3_-_36602069 | 8.77 |
ENSDART00000165414
|
rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr6_-_10725847 | 8.74 |
ENSDART00000184567
|
sp3b
|
Sp3b transcription factor |
| chr15_-_14642186 | 8.69 |
ENSDART00000164166
|
si:dkey-260j18.2
|
si:dkey-260j18.2 |
| chr13_+_9559461 | 8.66 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr23_-_7052362 | 8.55 |
ENSDART00000127702
ENSDART00000192468 |
prpf6
|
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) |
| chr7_-_30177691 | 8.53 |
ENSDART00000046689
|
tmed3
|
transmembrane p24 trafficking protein 3 |
| chr3_-_27065477 | 8.53 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr19_-_34742440 | 8.52 |
ENSDART00000122625
ENSDART00000175621 |
elp2
|
elongator acetyltransferase complex subunit 2 |
| chr7_+_38744924 | 8.49 |
ENSDART00000052329
|
znf408
|
zinc finger protein 408 |
| chr25_+_3549841 | 8.44 |
ENSDART00000164030
|
ccdc77
|
coiled-coil domain containing 77 |
| chr22_-_621888 | 8.42 |
ENSDART00000135829
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr5_+_51833132 | 8.41 |
ENSDART00000167491
|
papd4
|
PAP associated domain containing 4 |
| chr4_+_13931578 | 8.27 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr21_-_25213616 | 8.23 |
ENSDART00000122513
|
rfc2
|
replication factor C (activator 1) 2 |
| chr22_-_5655680 | 8.19 |
ENSDART00000159629
|
mcm2
|
minichromosome maintenance complex component 2 |
| chr9_-_12269847 | 8.19 |
ENSDART00000136558
ENSDART00000144734 ENSDART00000131766 ENSDART00000032344 |
nup35
|
nucleoporin 35 |
| chr20_-_37820939 | 8.16 |
ENSDART00000032978
|
nsl1
|
NSL1, MIS12 kinetochore complex component |
| chr8_+_387622 | 8.15 |
ENSDART00000167361
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr6_+_41808673 | 8.09 |
ENSDART00000038163
|
rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr5_+_68826514 | 8.06 |
ENSDART00000061406
|
usp39
|
ubiquitin specific peptidase 39 |
| chr6_+_49095646 | 7.97 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr13_-_24396199 | 7.90 |
ENSDART00000181093
|
tbp
|
TATA box binding protein |
| chr23_-_44871987 | 7.88 |
ENSDART00000085764
|
wu:fb72h05
|
wu:fb72h05 |
| chr7_-_16598212 | 7.83 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr3_-_61116258 | 7.81 |
ENSDART00000009194
ENSDART00000156978 |
aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr16_+_33931032 | 7.79 |
ENSDART00000167240
|
snip1
|
Smad nuclear interacting protein |
| chr19_+_7424347 | 7.79 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr5_-_4297459 | 7.77 |
ENSDART00000018895
|
srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr21_-_30994577 | 7.75 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr15_+_30350061 | 7.75 |
ENSDART00000135632
ENSDART00000044452 |
mrps23
|
mitochondrial ribosomal protein S23 |
| chr12_-_7253270 | 7.75 |
ENSDART00000035762
|
ube2d1b
|
ubiquitin-conjugating enzyme E2D 1b |
| chr12_+_20667301 | 7.75 |
ENSDART00000144804
|
mxra7
|
matrix-remodelling associated 7 |
| chr4_-_2265271 | 7.69 |
ENSDART00000125815
|
krr1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr21_-_45878872 | 7.57 |
ENSDART00000029763
|
sap30l
|
sap30-like |
| chr24_-_36196077 | 7.52 |
ENSDART00000154395
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr23_-_19715557 | 7.41 |
ENSDART00000143764
|
rpl10
|
ribosomal protein L10 |
| chr6_-_54433995 | 7.39 |
ENSDART00000017230
|
snrpc
|
small nuclear ribonucleoprotein polypeptide C |
| chr5_-_26893310 | 7.38 |
ENSDART00000126669
|
lman2lb
|
lectin, mannose-binding 2-like b |
| chr20_+_26892761 | 7.38 |
ENSDART00000133293
|
ftr97
|
finTRIM family, member 97 |
| chr1_+_34685405 | 7.36 |
ENSDART00000037986
ENSDART00000166007 |
gpalpp1
|
GPALPP motifs containing 1 |
| chr2_+_1124512 | 7.35 |
ENSDART00000108718
|
polr2d
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr13_-_24396003 | 7.34 |
ENSDART00000016211
|
tbp
|
TATA box binding protein |
| chr3_-_53559581 | 7.33 |
ENSDART00000183499
|
notch3
|
notch 3 |
| chr6_+_29707129 | 7.32 |
ENSDART00000023549
ENSDART00000164849 |
phb2b
|
prohibitin 2b |
| chr8_+_44613135 | 7.31 |
ENSDART00000063392
|
lsm1
|
LSM1, U6 small nuclear RNA associated |
| chr15_+_17343319 | 7.29 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr5_-_48070779 | 7.28 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr3_-_15444396 | 7.24 |
ENSDART00000104361
|
si:dkey-56d12.4
|
si:dkey-56d12.4 |
| chr18_-_35407289 | 7.21 |
ENSDART00000012018
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr7_-_13906409 | 7.20 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr13_+_24662238 | 7.19 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr17_+_45395846 | 7.13 |
ENSDART00000058793
|
nenf
|
neudesin neurotrophic factor |
| chr20_-_10487951 | 7.10 |
ENSDART00000064112
|
glrx5
|
glutaredoxin 5 homolog (S. cerevisiae) |
| chr1_-_44581937 | 7.09 |
ENSDART00000009858
|
tmx2b
|
thioredoxin-related transmembrane protein 2b |
| chr17_-_12966907 | 7.08 |
ENSDART00000022874
|
psma6a
|
proteasome subunit alpha 6a |
| chr7_+_22796723 | 7.05 |
ENSDART00000077395
ENSDART00000146368 |
rbm4.2
|
RNA binding motif protein 4.2 |
| chr12_-_20409794 | 6.98 |
ENSDART00000077936
|
lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr5_+_61862199 | 6.94 |
ENSDART00000132452
|
sympk
|
symplekin |
| chr21_+_22738939 | 6.90 |
ENSDART00000151342
ENSDART00000079145 |
arhgap42a
|
Rho GTPase activating protein 42a |
| chr12_-_13336703 | 6.86 |
ENSDART00000134356
|
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr21_+_6136981 | 6.86 |
ENSDART00000065859
ENSDART00000166287 |
cdk9
|
cyclin-dependent kinase 9 (CDC2-related kinase) |
| chr1_-_34685329 | 6.85 |
ENSDART00000125944
ENSDART00000008277 |
pibf1
|
progesterone immunomodulatory binding factor 1 |
| chr18_-_20458412 | 6.85 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr14_-_25935167 | 6.83 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr8_+_47677208 | 6.83 |
ENSDART00000123254
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr5_-_52010122 | 6.82 |
ENSDART00000073627
ENSDART00000163898 ENSDART00000051003 |
cdk7
|
cyclin-dependent kinase 7 |
| chr13_-_33654931 | 6.80 |
ENSDART00000020350
|
snx5
|
sorting nexin 5 |
| chr18_-_20458840 | 6.77 |
ENSDART00000177125
|
kif23
|
kinesin family member 23 |
| chr18_-_35407530 | 6.77 |
ENSDART00000137663
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr4_+_646154 | 6.73 |
ENSDART00000171138
|
xpot
|
exportin, tRNA (nuclear export receptor for tRNAs) |
| chr6_+_40775800 | 6.71 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr3_-_25275364 | 6.71 |
ENSDART00000163782
ENSDART00000145420 ENSDART00000133718 ENSDART00000055492 |
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr5_+_32490238 | 6.68 |
ENSDART00000191839
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr21_+_28502340 | 6.68 |
ENSDART00000077897
ENSDART00000140229 |
otub1a
|
OTU deubiquitinase, ubiquitin aldehyde binding 1a |
| chr3_-_34816893 | 6.68 |
ENSDART00000084448
ENSDART00000154696 |
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr8_+_15277874 | 6.67 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr18_+_26429428 | 6.66 |
ENSDART00000142686
|
blm
|
Bloom syndrome, RecQ helicase-like |
| chr8_+_47683539 | 6.66 |
ENSDART00000190701
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr23_+_37090889 | 6.64 |
ENSDART00000074406
|
ubxn10
|
UBX domain protein 10 |
| chr11_-_287670 | 6.62 |
ENSDART00000035737
|
slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr16_+_26680508 | 6.61 |
ENSDART00000142215
ENSDART00000159064 |
virma
|
vir like m6A methyltransferase associated |
| chr19_-_18855717 | 6.59 |
ENSDART00000158192
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr23_+_9560797 | 6.54 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr10_-_5135788 | 6.51 |
ENSDART00000108587
ENSDART00000138537 |
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr13_+_23897975 | 6.50 |
ENSDART00000002244
|
sf3b5
|
splicing factor 3b, subunit 5 |
| chr2_+_6243144 | 6.50 |
ENSDART00000058258
|
gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr13_-_35760969 | 6.49 |
ENSDART00000127476
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr7_+_1505507 | 6.48 |
ENSDART00000161015
|
nop10
|
NOP10 ribonucleoprotein homolog (yeast) |
| chr18_-_35407695 | 6.43 |
ENSDART00000191845
ENSDART00000141703 |
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr10_+_7671260 | 6.41 |
ENSDART00000157608
|
fam136a
|
family with sequence similarity 136, member A |
| chr4_-_18840919 | 6.39 |
ENSDART00000015834
|
cbll1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr8_+_41229233 | 6.38 |
ENSDART00000131135
|
zgc:152830
|
zgc:152830 |
| chr12_-_2869565 | 6.36 |
ENSDART00000152772
|
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr8_+_23726708 | 6.36 |
ENSDART00000142395
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr19_-_29853402 | 6.35 |
ENSDART00000024292
ENSDART00000188508 |
txlna
|
taxilin alpha |
| chr3_-_27066451 | 6.27 |
ENSDART00000156228
ENSDART00000156311 |
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr2_+_2772447 | 6.26 |
ENSDART00000124882
|
thoc1
|
THO complex 1 |
| chr23_-_14216506 | 6.26 |
ENSDART00000019620
|
ddx23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr5_-_25733745 | 6.25 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr7_+_71683853 | 6.17 |
ENSDART00000163002
|
emilin2b
|
elastin microfibril interfacer 2b |
| chr1_-_51720633 | 6.16 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr3_-_45777226 | 6.14 |
ENSDART00000192849
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr1_-_34685007 | 6.13 |
ENSDART00000157471
|
klf5a
|
Kruppel-like factor 5a |
| chr21_-_30181732 | 6.12 |
ENSDART00000015636
|
hnrnph1l
|
heterogeneous nuclear ribonucleoprotein H1, like |
| chr18_+_49248389 | 6.12 |
ENSDART00000059285
ENSDART00000142004 ENSDART00000132751 |
yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr22_+_9523479 | 6.10 |
ENSDART00000189473
ENSDART00000143953 |
strip1
|
striatin interacting protein 1 |
| chr14_-_26465729 | 6.07 |
ENSDART00000143454
|
syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr19_-_43657468 | 6.06 |
ENSDART00000150940
|
si:ch211-193k19.2
|
si:ch211-193k19.2 |
| chr1_+_25650917 | 6.05 |
ENSDART00000054235
|
plrg1
|
pleiotropic regulator 1 |
| chr18_-_21910991 | 6.04 |
ENSDART00000089787
ENSDART00000169220 ENSDART00000132381 ENSDART00000191764 |
edc4
|
enhancer of mRNA decapping 4 |
| chr10_-_3427589 | 6.04 |
ENSDART00000133452
ENSDART00000037183 |
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr20_-_29633507 | 6.03 |
ENSDART00000040292
|
cpsf3
|
cleavage and polyadenylation specific factor 3 |
| chr21_-_7781555 | 6.02 |
ENSDART00000084380
ENSDART00000189131 |
aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr2_-_5723786 | 6.01 |
ENSDART00000100924
|
cwc25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
| chr16_+_38337783 | 6.00 |
ENSDART00000135008
|
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
| chr20_-_16906623 | 5.96 |
ENSDART00000012859
ENSDART00000171628 |
psma6b
|
proteasome subunit alpha 6b |
Network of associatons between targets according to the STRING database.
First level regulatory network of etv5a+etv5b_elk1_elk4_etv1_elf1_erf+erfl3_gabpa_elk3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 31.5 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 5.9 | 17.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 4.6 | 13.8 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 4.2 | 12.6 | GO:1990403 | embryonic brain development(GO:1990403) |
| 3.6 | 10.8 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 3.5 | 24.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 3.3 | 9.9 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 3.3 | 13.2 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 3.0 | 18.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 2.9 | 8.8 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 2.8 | 19.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 2.6 | 10.6 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 2.5 | 7.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 2.4 | 19.1 | GO:0072422 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 2.4 | 11.8 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 2.3 | 11.6 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 2.2 | 6.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 2.2 | 33.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 2.2 | 15.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 2.0 | 30.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 2.0 | 5.9 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 1.9 | 5.8 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 1.8 | 12.4 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 1.8 | 12.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 1.8 | 8.8 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 1.8 | 8.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 1.7 | 6.9 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 1.7 | 5.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 1.7 | 25.6 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 1.7 | 6.8 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 1.7 | 6.8 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 1.7 | 5.0 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 1.7 | 6.7 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 1.7 | 16.6 | GO:0090329 | regulation of DNA-dependent DNA replication(GO:0090329) |
| 1.7 | 6.6 | GO:0010039 | response to iron ion(GO:0010039) |
| 1.6 | 4.9 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 1.6 | 8.1 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 1.6 | 6.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 1.6 | 1.6 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 1.6 | 4.7 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 1.5 | 13.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 1.5 | 4.6 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 1.5 | 6.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 1.5 | 11.9 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 1.4 | 10.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 1.4 | 11.6 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 1.4 | 4.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 1.4 | 4.3 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 1.4 | 5.6 | GO:0051645 | Golgi localization(GO:0051645) |
| 1.4 | 11.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 1.4 | 4.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 1.4 | 12.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 1.4 | 4.1 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 1.3 | 13.5 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 1.3 | 17.5 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 1.2 | 26.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 1.2 | 7.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 1.2 | 8.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 1.2 | 17.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 1.2 | 7.1 | GO:1901314 | regulation of histone ubiquitination(GO:0033182) negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 1.2 | 24.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 1.2 | 8.2 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 1.2 | 3.5 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 1.2 | 4.7 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 1.2 | 1.2 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 1.1 | 4.6 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 1.1 | 3.4 | GO:1903173 | fatty alcohol metabolic process(GO:1903173) |
| 1.1 | 2.2 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 1.1 | 4.4 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 1.1 | 4.4 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 1.1 | 27.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 1.0 | 3.1 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 1.0 | 6.0 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) histone mRNA metabolic process(GO:0008334) |
| 1.0 | 3.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 1.0 | 2.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 1.0 | 7.8 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 1.0 | 2.9 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 1.0 | 11.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 1.0 | 2.9 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.9 | 28.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.9 | 10.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.9 | 20.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.9 | 4.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.9 | 11.0 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.9 | 11.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.9 | 2.6 | GO:0071033 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.9 | 10.4 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.8 | 12.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.8 | 4.2 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.8 | 4.2 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.8 | 5.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.8 | 12.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.8 | 19.0 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.8 | 12.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.8 | 8.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.8 | 12.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.8 | 4.0 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.8 | 8.0 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.8 | 5.5 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.8 | 4.0 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.8 | 11.1 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.8 | 2.4 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.8 | 1.5 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.8 | 3.8 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.7 | 5.2 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.7 | 5.2 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.7 | 2.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.7 | 1.4 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.7 | 7.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.7 | 1.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.7 | 17.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.7 | 3.4 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.7 | 2.0 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.7 | 1.3 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.7 | 18.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.7 | 2.0 | GO:0030237 | female sex determination(GO:0030237) |
| 0.7 | 5.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.7 | 2.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.6 | 6.9 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.6 | 1.8 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.6 | 3.0 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.6 | 3.0 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.6 | 3.0 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.6 | 1.8 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.6 | 1.2 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.6 | 23.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.6 | 1.2 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.6 | 3.0 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.6 | 3.5 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.6 | 18.8 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.6 | 3.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.6 | 18.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.6 | 1.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.6 | 10.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.6 | 3.3 | GO:0046471 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.5 | 56.4 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.5 | 1.6 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.5 | 5.9 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.5 | 6.4 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.5 | 9.6 | GO:0050685 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.5 | 1.6 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.5 | 3.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.5 | 1.6 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.5 | 6.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.5 | 15.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.5 | 14.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.5 | 1.5 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.5 | 4.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.5 | 2.9 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.5 | 1.4 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.5 | 167.4 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.5 | 5.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.5 | 1.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.5 | 2.7 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.5 | 6.9 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.5 | 4.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.5 | 1.4 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.5 | 6.8 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.5 | 5.4 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.5 | 2.3 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.4 | 1.3 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.4 | 3.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 9.9 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.4 | 2.6 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.4 | 3.4 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.4 | 6.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.4 | 0.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.4 | 3.4 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.4 | 3.8 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.4 | 3.0 | GO:0031057 | negative regulation of histone modification(GO:0031057) |
| 0.4 | 1.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.4 | 7.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.4 | 1.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.4 | 4.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.4 | 0.8 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.4 | 7.9 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.4 | 21.3 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.4 | 0.4 | GO:1904035 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.4 | 1.2 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.4 | 4.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.4 | 6.8 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.4 | 7.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.4 | 1.9 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.4 | 1.5 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.4 | 3.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.4 | 1.5 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.4 | 1.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.4 | 8.3 | GO:1903322 | positive regulation of protein ubiquitination(GO:0031398) positive regulation of protein modification by small protein conjugation or removal(GO:1903322) |
| 0.4 | 4.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.4 | 1.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.4 | 0.7 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) |
| 0.4 | 2.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.3 | 1.7 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.3 | 13.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.3 | 1.4 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.3 | 5.1 | GO:0032392 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.3 | 4.0 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.3 | 0.7 | GO:0009188 | ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.3 | 1.7 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.3 | 3.6 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.3 | 2.3 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.3 | 23.6 | GO:0016072 | rRNA processing(GO:0006364) rRNA metabolic process(GO:0016072) |
| 0.3 | 1.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.3 | 1.6 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.3 | 1.3 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.3 | 2.5 | GO:0031123 | RNA 3'-end processing(GO:0031123) |
| 0.3 | 1.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.3 | 12.9 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.3 | 0.9 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.3 | 0.6 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.3 | 1.2 | GO:1903651 | positive regulation of cytoplasmic transport(GO:1903651) |
| 0.3 | 6.5 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.3 | 4.9 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.3 | 12.2 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.3 | 5.1 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.3 | 3.8 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.3 | 2.6 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.3 | 8.9 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.3 | 1.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.3 | 4.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.3 | 2.7 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.3 | 1.9 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.3 | 2.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.3 | 5.6 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.3 | 4.4 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.3 | 0.8 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.3 | 5.1 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.3 | 7.1 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.3 | 5.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.3 | 19.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.2 | 2.0 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 0.7 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 1.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 1.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.7 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.2 | 4.6 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 3.1 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 1.7 | GO:0010830 | regulation of myotube differentiation(GO:0010830) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 | 2.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 15.5 | GO:0018210 | peptidyl-threonine modification(GO:0018210) |
| 0.2 | 1.9 | GO:0036372 | opsin transport(GO:0036372) |
| 0.2 | 0.9 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.2 | 1.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 2.5 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.2 | 3.2 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.2 | 3.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 1.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 0.7 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 4.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 1.9 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.2 | 5.6 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.2 | 3.3 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.2 | 0.6 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 2.0 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.2 | 3.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 5.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.2 | 1.4 | GO:0070073 | regulation of melanocyte differentiation(GO:0045634) clustering of voltage-gated calcium channels(GO:0070073) |
| 0.2 | 0.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.2 | 7.9 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 2.6 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.2 | 10.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 3.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 1.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 0.6 | GO:0042560 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 4.9 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.2 | 3.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 0.7 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 4.8 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 2.3 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.2 | 2.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 15.3 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.2 | 1.4 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.2 | 2.0 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.2 | 9.5 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.2 | 1.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.2 | 3.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.2 | 0.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.2 | 3.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 0.5 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.2 | 1.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 3.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 0.8 | GO:0071422 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.2 | 0.6 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.2 | 0.9 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 0.8 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.2 | 3.7 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.2 | 2.4 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 0.7 | GO:0034982 | protein processing involved in protein targeting to mitochondrion(GO:0006627) mitochondrial protein processing(GO:0034982) |
| 0.1 | 0.7 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.7 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.4 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 1.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 5.1 | GO:0033500 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.1 | 0.9 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 0.6 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.7 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 6.4 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 5.5 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 0.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.3 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 1.7 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.1 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.4 | GO:0035844 | cloaca development(GO:0035844) |
| 0.1 | 0.9 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 3.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 4.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.8 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 1.7 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.1 | 0.5 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 3.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 2.9 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 0.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.6 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 3.0 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.1 | 2.6 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.1 | 6.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 8.3 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 0.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 1.2 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 2.8 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 1.3 | GO:0014074 | response to purine-containing compound(GO:0014074) response to ATP(GO:0033198) response to organophosphorus(GO:0046683) |
| 0.1 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 2.1 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.6 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 1.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.2 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.1 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 3.1 | GO:0060840 | artery development(GO:0060840) |
| 0.1 | 6.3 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.1 | 7.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 1.5 | GO:0098781 | ncRNA transcription(GO:0098781) |
| 0.1 | 0.8 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 0.9 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 10.0 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.1 | 1.9 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell development(GO:0072015) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell development(GO:0072310) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.5 | GO:0003308 | desmosome assembly(GO:0002159) negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.1 | 0.3 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.1 | 1.1 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 1.8 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.1 | 6.2 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.1 | 1.3 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 2.9 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.1 | 1.5 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 4.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.1 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 0.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 5.3 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 2.4 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.1 | 2.0 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 0.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.1 | GO:0033003 | regulation of mast cell activation(GO:0033003) |
| 0.1 | 0.2 | GO:0051125 | regulation of actin nucleation(GO:0051125) |
| 0.1 | 0.3 | GO:0043901 | negative regulation of multi-organism process(GO:0043901) negative regulation of oocyte development(GO:0060283) |
| 0.1 | 0.6 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 1.1 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 1.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 3.4 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 5.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 1.3 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) frontonasal suture morphogenesis(GO:0097095) |
| 0.1 | 1.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 3.3 | GO:0009306 | protein secretion(GO:0009306) |
| 0.1 | 2.9 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 0.3 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 12.4 | GO:0061371 | determination of heart left/right asymmetry(GO:0061371) |
| 0.1 | 1.5 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.1 | 1.1 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 2.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.8 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 3.0 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 3.7 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 0.6 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.3 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.1 | 5.5 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 3.7 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 3.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 0.9 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 7.9 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.1 | 3.3 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.1 | 1.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 3.3 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 | 2.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 6.6 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.1 | 2.3 | GO:0048565 | digestive tract development(GO:0048565) |
| 0.1 | 0.7 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.1 | 0.8 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.2 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.1 | 3.0 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 4.6 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.1 | 2.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 3.5 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 1.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.8 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.1 | 0.5 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.7 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.3 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 7.1 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 1.5 | GO:1903706 | regulation of hemopoiesis(GO:1903706) |
| 0.0 | 1.2 | GO:0009145 | purine nucleoside triphosphate biosynthetic process(GO:0009145) purine ribonucleoside triphosphate biosynthetic process(GO:0009206) |
| 0.0 | 0.5 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 1.0 | GO:0051961 | negative regulation of neurogenesis(GO:0050768) negative regulation of nervous system development(GO:0051961) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 20.1 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.0 | 0.9 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.8 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 2.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.7 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 2.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 6.3 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.4 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 1.9 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 0.4 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 2.5 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.0 | 1.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0060035 | notochord cell development(GO:0060035) notochord cell vacuolation(GO:0060036) |
| 0.0 | 1.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.0 | 0.9 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.5 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 4.2 | GO:0022604 | regulation of cell morphogenesis(GO:0022604) |
| 0.0 | 0.3 | GO:0034284 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 2.4 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.3 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.1 | GO:2001057 | reactive nitrogen species metabolic process(GO:2001057) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.1 | GO:0009953 | dorsal/ventral pattern formation(GO:0009953) |
| 0.0 | 0.9 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 0.2 | GO:0071453 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0030593 | neutrophil chemotaxis(GO:0030593) granulocyte chemotaxis(GO:0071621) |
| 0.0 | 0.4 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.2 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.4 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.2 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0030814 | regulation of cyclic nucleotide metabolic process(GO:0030799) regulation of cyclic nucleotide biosynthetic process(GO:0030802) regulation of nucleotide biosynthetic process(GO:0030808) regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of cyclase activity(GO:0031279) regulation of adenylate cyclase activity(GO:0045761) regulation of lyase activity(GO:0051339) regulation of purine nucleotide biosynthetic process(GO:1900371) |
| 0.0 | 0.6 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 1.1 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.4 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 0.2 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.1 | GO:0006284 | base-excision repair(GO:0006284) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 4.9 | 34.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 4.4 | 17.6 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 3.5 | 10.6 | GO:0098536 | deuterosome(GO:0098536) |
| 3.5 | 13.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 2.8 | 11.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 2.7 | 13.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 2.6 | 28.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 2.5 | 12.4 | GO:0031415 | NatA complex(GO:0031415) |
| 2.3 | 9.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 2.2 | 13.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 2.0 | 10.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 2.0 | 18.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 1.9 | 9.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 1.8 | 42.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 1.8 | 9.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 1.7 | 43.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 1.5 | 6.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.5 | 10.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.5 | 16.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.5 | 16.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.5 | 9.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.5 | 10.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 1.4 | 4.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 1.3 | 14.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 1.3 | 9.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 1.3 | 7.6 | GO:0070449 | elongin complex(GO:0070449) |
| 1.3 | 17.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 1.2 | 19.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 1.2 | 15.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 1.1 | 33.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 1.1 | 15.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 1.1 | 3.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 1.0 | 9.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 1.0 | 11.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 1.0 | 10.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 1.0 | 6.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 1.0 | 19.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.9 | 5.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.9 | 6.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.9 | 2.7 | GO:0034708 | methyltransferase complex(GO:0034708) |
| 0.9 | 15.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.9 | 14.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.9 | 8.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.9 | 20.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.9 | 4.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.8 | 7.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.8 | 11.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.8 | 4.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.8 | 17.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.8 | 12.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.8 | 10.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.7 | 8.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.7 | 4.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.7 | 15.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.7 | 2.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.7 | 6.8 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.7 | 2.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.7 | 2.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.7 | 5.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.6 | 7.8 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.6 | 14.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.6 | 3.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.6 | 1.8 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.6 | 4.7 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.6 | 1.8 | GO:0097361 | CIA complex(GO:0097361) |
| 0.6 | 4.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.6 | 11.6 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.6 | 2.9 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.6 | 2.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.6 | 10.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.6 | 4.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.6 | 3.4 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.6 | 3.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.6 | 6.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.5 | 7.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.5 | 4.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.5 | 4.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.5 | 3.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.5 | 2.6 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.5 | 2.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.5 | 1.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.5 | 3.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.5 | 4.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.5 | 8.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.5 | 5.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.5 | 0.5 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.5 | 1.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.5 | 36.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.5 | 11.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.4 | 3.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.4 | 18.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 4.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 1.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 0.4 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.4 | 28.4 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.4 | 2.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.4 | 2.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 3.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.4 | 16.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.4 | 3.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.4 | 18.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.4 | 11.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.4 | 7.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.4 | 1.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.4 | 37.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.4 | 11.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 7.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 2.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.3 | 1.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.3 | 5.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 25.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.3 | 3.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 1.9 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 1.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.3 | 5.2 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.3 | 1.5 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.3 | 2.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 18.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.3 | 5.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 2.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.3 | 4.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.3 | 0.3 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.3 | 7.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 3.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 1.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.3 | 1.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.3 | 0.8 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.3 | 1.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.3 | 3.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.3 | 1.9 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.3 | 58.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.3 | 14.0 | GO:0016604 | nuclear body(GO:0016604) |
| 0.3 | 13.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 0.8 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.3 | 2.0 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.2 | 3.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 3.6 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.2 | 1.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.2 | 2.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 3.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 2.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 3.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 3.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 1.5 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.2 | 0.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 5.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 0.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 0.7 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 2.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.2 | 1.4 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.2 | 4.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 30.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.2 | 1.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 14.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 0.8 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 5.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.2 | 0.8 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 0.3 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 1.6 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 4.0 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 3.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 41.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 1.0 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.7 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 2.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 2.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 9.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.7 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 8.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 10.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 17.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 4.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 4.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.5 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 2.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 4.1 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.1 | 1.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 10.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 5.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 11.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 6.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.7 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 9.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 1.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 342.2 | GO:0005634 | nucleus(GO:0005634) |
| 0.1 | 0.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 3.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 1.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 1.8 | GO:0045111 | intermediate filament(GO:0005882) intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 1.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.5 | 31.5 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 5.9 | 17.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 4.8 | 14.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 4.6 | 13.8 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 4.1 | 32.4 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 3.1 | 12.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 2.9 | 11.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 2.6 | 10.6 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 2.5 | 9.9 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 2.3 | 27.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 2.2 | 11.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 2.2 | 6.6 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 2.2 | 6.5 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 1.9 | 21.3 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 1.9 | 5.8 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 1.9 | 11.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.7 | 6.7 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 1.7 | 13.2 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 1.5 | 16.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 1.4 | 7.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.4 | 12.4 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 1.4 | 15.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 1.3 | 5.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 1.3 | 2.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.2 | 3.7 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 1.2 | 2.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 1.1 | 8.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 1.1 | 8.8 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 1.1 | 10.6 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.0 | 9.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.0 | 3.1 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 1.0 | 15.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 1.0 | 12.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 1.0 | 17.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 1.0 | 8.7 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 1.0 | 3.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 1.0 | 4.8 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 1.0 | 3.8 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.9 | 7.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.9 | 4.7 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.9 | 2.8 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.9 | 2.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.9 | 6.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.9 | 8.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.9 | 5.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.9 | 7.7 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.9 | 19.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.8 | 8.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.8 | 5.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.8 | 21.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.8 | 4.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.8 | 12.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.8 | 4.0 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.8 | 10.3 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.8 | 4.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.8 | 4.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.8 | 2.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.7 | 3.7 | GO:0008106 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.7 | 3.7 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.7 | 3.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.7 | 37.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.7 | 14.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.7 | 15.0 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.7 | 5.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.7 | 12.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.7 | 3.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.7 | 5.5 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.7 | 2.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.7 | 8.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.7 | 18.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.7 | 8.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.7 | 3.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.7 | 3.3 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.6 | 1.9 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.6 | 4.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.6 | 3.0 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.6 | 1.8 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.6 | 10.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.6 | 3.5 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.6 | 1.8 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.6 | 2.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.6 | 15.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.6 | 1.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.6 | 1.7 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.6 | 5.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.5 | 9.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.5 | 3.7 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.5 | 2.6 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.5 | 12.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.5 | 1.0 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.5 | 6.0 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.5 | 2.0 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.5 | 0.9 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.5 | 3.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.4 | 1.3 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.4 | 5.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.4 | 3.8 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.4 | 5.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.4 | 34.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.4 | 5.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.4 | 2.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.4 | 1.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.4 | 2.8 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.4 | 7.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.4 | 14.9 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.4 | 3.7 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.4 | 3.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 1.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 11.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.3 | 2.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 5.7 | GO:0004532 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.3 | 1.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 3.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 2.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.3 | 1.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.3 | 2.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 4.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.3 | 2.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 7.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.3 | 1.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.3 | 1.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 16.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.3 | 2.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.3 | 14.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.3 | 2.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 17.0 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.3 | 2.3 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 4.6 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.3 | 5.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.3 | 21.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.3 | 1.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.3 | 1.7 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.3 | 7.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.3 | 1.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.3 | 1.1 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.3 | 0.8 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 1.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 1.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 6.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.3 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 11.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.3 | 1.0 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 4.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.7 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 1.7 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 12.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 0.7 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.2 | 0.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 0.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 7.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 4.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 3.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 1.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 3.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 3.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 9.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 0.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 16.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.2 | 0.6 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.2 | 15.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 0.6 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 1.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 1.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 2.9 | GO:0002020 | protease binding(GO:0002020) |
| 0.2 | 3.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 10.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 6.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.2 | 4.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 170.7 | GO:0003723 | RNA binding(GO:0003723) |
| 0.2 | 12.8 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.2 | 1.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 2.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 0.2 | GO:0052813 | phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 1.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 1.5 | GO:0015154 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 0.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 21.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 0.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 5.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 5.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 10.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 3.9 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 4.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.2 | 1.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 0.5 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.2 | 3.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 1.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 0.8 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 5.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 2.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 2.7 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 1.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 2.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 2.1 | GO:0061608 | nuclear localization sequence binding(GO:0008139) nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 2.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 1.7 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 2.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 2.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 2.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.8 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 46.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 3.7 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 0.5 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 1.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 2.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 12.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.2 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 1.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 6.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 1.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 14.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 39.5 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.1 | 18.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 4.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.3 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.1 | 7.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.5 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 2.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 24.9 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.1 | 1.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 8.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 5.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 1.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 1.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 21.9 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.1 | 1.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 1.6 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 1.6 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 0.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 1.0 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 14.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.8 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.5 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.1 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 0.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 28.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 1.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 2.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.2 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.0 | 0.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 22.2 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.7 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 1.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.3 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 14.0 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 13.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 7.5 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
| 0.0 | 90.6 | GO:0003676 | nucleic acid binding(GO:0003676) |
| 0.0 | 0.3 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 3.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 6.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 10.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.6 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleoside transmembrane transporter activity(GO:0015211) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.0 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 1.6 | 1.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 1.1 | 15.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.9 | 9.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.9 | 1.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.8 | 30.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.7 | 10.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.6 | 26.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.6 | 4.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.6 | 12.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 7.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.4 | 8.6 | PID ATM PATHWAY | ATM pathway |
| 0.4 | 20.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.4 | 5.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 6.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 8.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 2.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 2.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 16.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.3 | 16.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 0.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 10.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.3 | 7.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 1.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 9.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 15.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 7.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 2.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 3.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 2.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 2.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 3.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 4.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 0.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 13.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 3.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 0.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 5.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.5 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 2.5 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 2.4 | 14.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 1.9 | 33.0 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 1.6 | 63.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 1.5 | 36.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 1.4 | 10.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 1.3 | 44.6 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 1.3 | 13.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 1.2 | 33.6 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 1.2 | 22.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 1.2 | 18.7 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 1.1 | 19.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 1.0 | 20.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.9 | 11.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.9 | 12.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.8 | 26.6 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.8 | 17.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.8 | 12.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.8 | 4.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.7 | 19.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.7 | 5.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.7 | 6.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.7 | 19.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.7 | 13.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.6 | 13.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 35.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.6 | 9.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.6 | 5.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.6 | 4.6 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.5 | 5.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.5 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.4 | 32.4 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.4 | 4.7 | REACTOME DEADENYLATION DEPENDENT MRNA DECAY | Genes involved in Deadenylation-dependent mRNA decay |
| 0.4 | 6.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 6.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.4 | 6.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.4 | 3.0 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.3 | 2.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 13.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.3 | 11.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.3 | 9.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.3 | 4.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 4.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 4.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 5.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.3 | 25.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.3 | 6.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.3 | 3.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 3.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 14.1 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.2 | 2.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.2 | 2.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 11.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 2.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 1.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 1.6 | REACTOME METABOLISM OF RNA | Genes involved in Metabolism of RNA |
| 0.2 | 9.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.2 | 3.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 3.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 2.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 2.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 2.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 3.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.7 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 4.1 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 0.3 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.1 | 0.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.0 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.1 | 2.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 2.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.7 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 2.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |