Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
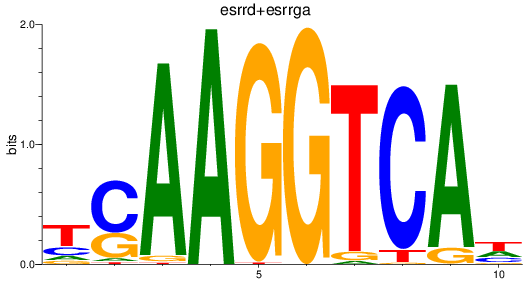
Results for esrrd+esrrga_esrrb_esrra+esrrgb
Z-value: 2.55
Transcription factors associated with esrrd+esrrga_esrrb_esrra+esrrgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
esrrga
|
ENSDARG00000004861 | estrogen-related receptor gamma a |
|
esrrd
|
ENSDARG00000015064 | estrogen-related receptor delta |
|
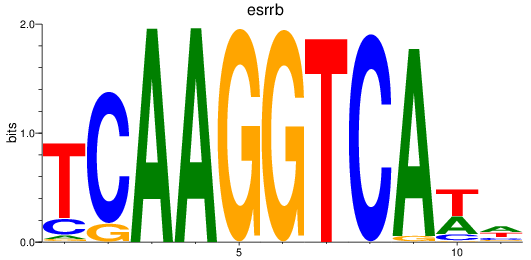
esrrb
|
ENSDARG00000100847 | estrogen-related receptor beta |
|
esrrgb
|
ENSDARG00000011696 | estrogen-related receptor gamma b |
|
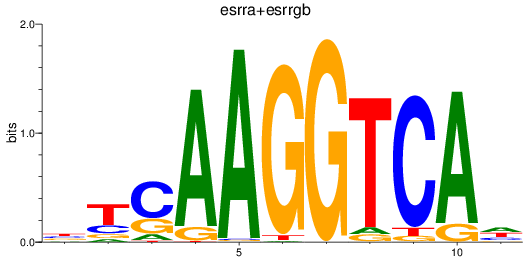
esrra
|
ENSDARG00000069266 | estrogen-related receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| esrrb | dr11_v1_chr17_-_29902187_29902187 | 0.81 | 2.3e-05 | Click! |
| esrra | dr11_v1_chr21_+_26612777_26612777 | 0.76 | 1.8e-04 | Click! |
| esrrgb | dr11_v1_chr20_-_46808870_46808870 | 0.71 | 6.3e-04 | Click! |
| esrrd | dr11_v1_chr18_-_48992363_48992363 | 0.57 | 1.1e-02 | Click! |
Activity profile of esrrd+esrrga_esrrb_esrra+esrrgb motif
Sorted Z-values of esrrd+esrrga_esrrb_esrra+esrrgb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_11385155 | 13.85 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr12_+_34896956 | 13.41 |
ENSDART00000055415
|
prph2a
|
peripherin 2a (retinal degeneration, slow) |
| chr9_+_22003942 | 12.30 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr7_+_35068036 | 10.58 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr17_-_12385308 | 10.02 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr13_+_50375800 | 9.95 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr15_+_20403903 | 9.80 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr9_-_44939104 | 8.62 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr5_-_37886063 | 8.38 |
ENSDART00000131378
ENSDART00000132152 |
si:ch211-139a5.9
|
si:ch211-139a5.9 |
| chr16_+_52512025 | 7.54 |
ENSDART00000056095
|
fabp10a
|
fatty acid binding protein 10a, liver basic |
| chr8_+_24854600 | 7.31 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr11_+_25472758 | 7.24 |
ENSDART00000011178
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr5_+_1278092 | 7.16 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr6_+_48618512 | 6.91 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr25_+_3326885 | 6.90 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr23_+_44611864 | 6.79 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr9_-_48736388 | 6.41 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr3_-_21280373 | 6.26 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr11_+_21076872 | 6.05 |
ENSDART00000155521
|
prelp
|
proline/arginine-rich end leucine-rich repeat protein |
| chr11_-_7320211 | 6.04 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr3_-_3209432 | 5.97 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr15_+_19797918 | 5.89 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr12_-_30841679 | 5.85 |
ENSDART00000105594
|
crygmx
|
crystallin, gamma MX |
| chr4_-_1360495 | 5.70 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr7_-_48805181 | 5.37 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr14_-_33454595 | 5.36 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr3_-_60142530 | 5.33 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr20_+_6142433 | 5.32 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr25_-_19648154 | 5.18 |
ENSDART00000148570
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr5_-_57820873 | 5.18 |
ENSDART00000089961
|
sik2a
|
salt-inducible kinase 2a |
| chr7_+_67494107 | 5.08 |
ENSDART00000185653
|
cpne7
|
copine VII |
| chr25_+_3327071 | 4.85 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr16_+_26012569 | 4.83 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr9_-_42861080 | 4.79 |
ENSDART00000193688
|
ttn.1
|
titin, tandem duplicate 1 |
| chr4_+_47736069 | 4.79 |
ENSDART00000193329
|
si:ch211-196f19.1
|
si:ch211-196f19.1 |
| chr21_-_32487061 | 4.71 |
ENSDART00000114359
ENSDART00000131591 ENSDART00000131477 |
si:dkeyp-72g9.4
|
si:dkeyp-72g9.4 |
| chr7_+_568819 | 4.70 |
ENSDART00000173716
|
nrxn2b
|
neurexin 2b |
| chr10_-_42685512 | 4.63 |
ENSDART00000081347
|
stc1l
|
stanniocalcin 1, like |
| chr19_+_9344171 | 4.60 |
ENSDART00000133447
ENSDART00000104622 |
si:ch211-288g17.4
|
si:ch211-288g17.4 |
| chr17_-_25737452 | 4.58 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr3_+_16762483 | 4.57 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr20_+_43942278 | 4.57 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr10_-_32524771 | 4.51 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr5_-_10768258 | 4.51 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr6_-_28980756 | 4.51 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr21_+_19008168 | 4.50 |
ENSDART00000136196
ENSDART00000128381 ENSDART00000176624 |
nefla
|
neurofilament, light polypeptide a |
| chr8_-_21372446 | 4.43 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr12_+_45200744 | 4.42 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr21_+_30563115 | 4.37 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr11_+_30513656 | 4.35 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr21_+_25231160 | 4.34 |
ENSDART00000063089
ENSDART00000139127 |
gng8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr8_-_44015210 | 4.33 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr2_-_58201173 | 4.22 |
ENSDART00000166282
|
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr5_+_21891305 | 4.19 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr20_-_1151265 | 4.19 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr19_+_20724347 | 4.18 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr4_+_5531583 | 4.17 |
ENSDART00000137500
ENSDART00000042080 |
si:dkey-14d8.6
|
si:dkey-14d8.6 |
| chr5_-_69934558 | 4.07 |
ENSDART00000124954
|
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr10_+_10677697 | 4.00 |
ENSDART00000188705
|
fam163b
|
family with sequence similarity 163, member B |
| chr22_+_19290199 | 3.95 |
ENSDART00000148173
|
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr18_+_910992 | 3.89 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr24_-_17023392 | 3.88 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr6_-_35439406 | 3.81 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr17_-_45254585 | 3.78 |
ENSDART00000185507
ENSDART00000172080 |
ttbk2a
|
tau tubulin kinase 2a |
| chr1_+_45080897 | 3.76 |
ENSDART00000129819
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr2_-_4797512 | 3.76 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr17_+_33340675 | 3.71 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr8_-_14052349 | 3.63 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr15_+_44472356 | 3.63 |
ENSDART00000155763
ENSDART00000028011 |
msantd4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr10_+_1638876 | 3.63 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr20_-_27857676 | 3.58 |
ENSDART00000192934
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr6_+_51773873 | 3.53 |
ENSDART00000156516
|
tmem74b
|
transmembrane protein 74B |
| chr2_-_19109304 | 3.50 |
ENSDART00000168028
|
si:dkey-225f23.5
|
si:dkey-225f23.5 |
| chr2_-_24289641 | 3.50 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr6_+_2190214 | 3.46 |
ENSDART00000156716
|
acvr1bb
|
activin A receptor type 1Bb |
| chr23_-_45504991 | 3.46 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr22_+_19289970 | 3.45 |
ENSDART00000137976
ENSDART00000132386 |
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr11_+_28476298 | 3.43 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr22_+_19188809 | 3.41 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr15_+_45563656 | 3.40 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr4_+_5537101 | 3.31 |
ENSDART00000008692
|
si:dkey-14d8.7
|
si:dkey-14d8.7 |
| chr4_+_17280868 | 3.29 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr22_-_21150845 | 3.29 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr25_+_7982979 | 3.29 |
ENSDART00000171904
|
ucmab
|
upper zone of growth plate and cartilage matrix associated b |
| chr11_-_29650930 | 3.27 |
ENSDART00000166969
|
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr1_-_54718863 | 3.21 |
ENSDART00000122601
|
pgam1b
|
phosphoglycerate mutase 1b |
| chr19_-_5103313 | 3.16 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr7_+_58699900 | 3.08 |
ENSDART00000144009
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr3_+_20012891 | 3.08 |
ENSDART00000078974
|
tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr1_+_44826593 | 3.08 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr24_+_32472155 | 3.07 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr20_+_5564042 | 3.06 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr6_+_52235441 | 3.06 |
ENSDART00000056319
|
cox6c
|
cytochrome c oxidase subunit VIc |
| chr2_+_53359234 | 3.06 |
ENSDART00000147581
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr6_+_40714811 | 3.04 |
ENSDART00000153868
|
ccdc36
|
coiled-coil domain containing 36 |
| chr24_+_33802528 | 3.03 |
ENSDART00000136040
ENSDART00000147499 ENSDART00000182322 |
atg9b
|
autophagy related 9B |
| chr25_-_35599887 | 3.03 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr15_+_45994123 | 2.99 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr4_+_2620751 | 2.99 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr6_+_46481024 | 2.98 |
ENSDART00000155596
|
si:dkey-7k24.5
|
si:dkey-7k24.5 |
| chr19_+_47311020 | 2.94 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr22_+_33362552 | 2.93 |
ENSDART00000101580
|
nicn1
|
nicolin 1 |
| chr6_-_607063 | 2.92 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr24_+_37080771 | 2.88 |
ENSDART00000159942
|
kcnc3b
|
potassium voltage-gated channel, Shaw-related subfamily, member 3b |
| chr11_-_10770053 | 2.88 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr10_+_45128375 | 2.88 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr7_-_16205471 | 2.88 |
ENSDART00000173584
|
btr05
|
bloodthirsty-related gene family, member 5 |
| chr14_-_9199968 | 2.88 |
ENSDART00000146113
|
arhgef9b
|
Cdc42 guanine nucleotide exchange factor (GEF) 9b |
| chr12_-_45304971 | 2.87 |
ENSDART00000186537
ENSDART00000126405 |
fdxr
|
ferredoxin reductase |
| chr2_+_37110504 | 2.85 |
ENSDART00000132794
ENSDART00000042974 |
slc1a8b
|
solute carrier family 1 (glutamate transporter), member 8b |
| chr7_-_38638276 | 2.84 |
ENSDART00000074463
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr10_-_19523752 | 2.82 |
ENSDART00000160057
|
si:ch211-127i16.2
|
si:ch211-127i16.2 |
| chr1_-_1627487 | 2.82 |
ENSDART00000166094
|
clic6
|
chloride intracellular channel 6 |
| chr21_+_25226558 | 2.81 |
ENSDART00000168480
|
sycn.2
|
syncollin, tandem duplicate 2 |
| chr3_+_61185660 | 2.78 |
ENSDART00000167114
|
CR352263.1
|
|
| chr15_-_7598542 | 2.77 |
ENSDART00000173092
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr10_+_21737745 | 2.77 |
ENSDART00000170498
ENSDART00000167997 |
pcdh1g18
|
protocadherin 1 gamma 18 |
| chr19_-_5103141 | 2.75 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr15_-_7598294 | 2.74 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr2_+_10134345 | 2.73 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr3_+_4997545 | 2.73 |
ENSDART00000181237
|
CABZ01117706.1
|
|
| chr17_-_12389259 | 2.72 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr9_+_7909490 | 2.70 |
ENSDART00000133063
ENSDART00000109288 |
myo16
|
myosin XVI |
| chr24_-_40009446 | 2.68 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr11_-_25213651 | 2.66 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr5_+_5689476 | 2.63 |
ENSDART00000022729
|
unm_sa808
|
un-named sa808 |
| chr3_+_43460696 | 2.63 |
ENSDART00000164581
|
galr2b
|
galanin receptor 2b |
| chr2_-_9646857 | 2.62 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr20_-_34090740 | 2.62 |
ENSDART00000062539
ENSDART00000008140 |
pdcb
|
phosducin b |
| chr3_+_1182315 | 2.61 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr17_-_38887424 | 2.60 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr16_-_13281380 | 2.60 |
ENSDART00000103882
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr11_-_11471857 | 2.60 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr6_-_10320676 | 2.60 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr5_+_69981453 | 2.56 |
ENSDART00000143860
|
si:ch211-154e10.1
|
si:ch211-154e10.1 |
| chr10_+_35952532 | 2.55 |
ENSDART00000184730
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr1_-_50859053 | 2.55 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr19_-_38872650 | 2.55 |
ENSDART00000146641
|
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr5_-_42544522 | 2.53 |
ENSDART00000157968
|
BX510949.1
|
|
| chr3_+_42923275 | 2.53 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr21_+_5882300 | 2.53 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr25_+_5604512 | 2.52 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr3_-_34724879 | 2.51 |
ENSDART00000177021
|
thraa
|
thyroid hormone receptor alpha a |
| chr10_-_5814101 | 2.50 |
ENSDART00000161903
|
il6st
|
interleukin 6 signal transducer |
| chr8_-_39903803 | 2.49 |
ENSDART00000012391
|
cabp1a
|
calcium binding protein 1a |
| chr7_+_48761875 | 2.48 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr11_+_36989696 | 2.46 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr3_-_28828569 | 2.46 |
ENSDART00000167679
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr20_+_49119633 | 2.44 |
ENSDART00000151435
|
cd109
|
CD109 molecule |
| chr7_-_50633285 | 2.43 |
ENSDART00000174018
|
crtc3
|
CREB regulated transcription coactivator 3 |
| chr25_+_25124684 | 2.43 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr4_-_11163112 | 2.40 |
ENSDART00000188854
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr9_-_52678146 | 2.40 |
ENSDART00000169179
|
pfkla
|
phosphofructokinase, liver a |
| chr6_-_10835849 | 2.40 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr19_-_5805923 | 2.38 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr21_+_25221940 | 2.37 |
ENSDART00000108972
|
sycn.1
|
syncollin, tandem duplicate 1 |
| chr16_-_45058919 | 2.32 |
ENSDART00000177134
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr8_-_39760427 | 2.31 |
ENSDART00000184336
ENSDART00000186500 |
si:ch211-170d8.8
|
si:ch211-170d8.8 |
| chr7_+_58699718 | 2.30 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr5_-_38064065 | 2.26 |
ENSDART00000137181
|
si:dkey-111e8.5
|
si:dkey-111e8.5 |
| chr7_+_9904627 | 2.25 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr5_-_57879138 | 2.25 |
ENSDART00000145959
|
sik2a
|
salt-inducible kinase 2a |
| chr3_-_41795917 | 2.20 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr1_-_55810730 | 2.18 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr23_+_6272638 | 2.16 |
ENSDART00000190366
|
syt2a
|
synaptotagmin IIa |
| chr18_-_226800 | 2.16 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr7_+_58686860 | 2.14 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr8_-_49908978 | 2.14 |
ENSDART00000172642
|
agtpbp1
|
ATP/GTP binding protein 1 |
| chr23_+_42434348 | 2.14 |
ENSDART00000161027
|
cyp2aa1
|
cytochrome P450, family 2, subfamily AA, polypeptide 1 |
| chr13_-_20540790 | 2.13 |
ENSDART00000131467
|
si:ch1073-126c3.2
|
si:ch1073-126c3.2 |
| chr5_-_15524782 | 2.13 |
ENSDART00000189781
|
CU694999.1
|
|
| chr16_+_11724230 | 2.12 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr2_-_8102169 | 2.12 |
ENSDART00000131955
|
pls1
|
plastin 1 (I isoform) |
| chr4_-_11165611 | 2.12 |
ENSDART00000059489
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr12_+_47162761 | 2.11 |
ENSDART00000192339
ENSDART00000167726 |
RYR2
|
ryanodine receptor 2 |
| chr4_-_11165961 | 2.10 |
ENSDART00000193393
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr17_-_29192987 | 2.07 |
ENSDART00000164302
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr17_-_51202339 | 2.07 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr20_-_39271844 | 2.05 |
ENSDART00000192708
|
clu
|
clusterin |
| chr5_-_37103487 | 2.05 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr3_-_32337653 | 2.05 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr5_+_28830388 | 2.03 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr22_+_7742211 | 2.02 |
ENSDART00000140896
|
CELA1 (1 of many)
|
zgc:92511 |
| chr5_-_24044877 | 2.01 |
ENSDART00000161131
|
pdha1a
|
pyruvate dehydrogenase E1 alpha 1 subunit a |
| chr16_-_50181206 | 2.01 |
ENSDART00000148478
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr9_-_42873700 | 2.01 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr22_+_9472814 | 1.99 |
ENSDART00000112125
ENSDART00000138850 |
cacna2d2b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2b |
| chr20_+_1412193 | 1.99 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr5_-_35301800 | 1.97 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr2_+_55982300 | 1.97 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr7_+_9981757 | 1.95 |
ENSDART00000113429
ENSDART00000173233 |
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr17_-_33289304 | 1.95 |
ENSDART00000135118
ENSDART00000040346 |
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr1_-_50838160 | 1.95 |
ENSDART00000163939
ENSDART00000165111 |
zgc:154142
|
zgc:154142 |
| chr8_-_39739056 | 1.95 |
ENSDART00000147992
|
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr25_+_31238606 | 1.95 |
ENSDART00000149439
|
tnni2a.2
|
troponin I type 2a (skeletal, fast), tandem duplicate 2 |
| chr5_-_69314495 | 1.93 |
ENSDART00000182335
|
smtnb
|
smoothelin b |
| chr10_+_22782522 | 1.92 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr11_-_18791834 | 1.92 |
ENSDART00000156431
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr12_+_47162456 | 1.91 |
ENSDART00000186272
ENSDART00000180811 |
RYR2
|
ryanodine receptor 2 |
| chr20_-_1141722 | 1.90 |
ENSDART00000152675
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of esrrd+esrrga_esrrb_esrra+esrrgb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0015824 | proline transport(GO:0015824) |
| 2.2 | 8.6 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 1.5 | 6.1 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 1.5 | 5.9 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.5 | 4.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 1.4 | 5.6 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.4 | 4.2 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 1.4 | 12.4 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 1.2 | 3.7 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 1.0 | 12.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.9 | 12.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.9 | 11.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.8 | 6.8 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.8 | 3.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.8 | 1.6 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.7 | 3.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.7 | 4.4 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.7 | 2.2 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.6 | 1.2 | GO:0051196 | regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) regulation of cofactor metabolic process(GO:0051193) regulation of coenzyme metabolic process(GO:0051196) |
| 0.6 | 3.5 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.5 | 2.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.5 | 6.2 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.5 | 2.5 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.4 | 2.6 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.4 | 1.3 | GO:0009750 | response to fructose(GO:0009750) |
| 0.4 | 2.6 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.4 | 6.8 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.4 | 1.3 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.4 | 1.3 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.4 | 1.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.4 | 4.5 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.4 | 1.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.4 | 3.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.4 | 11.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.4 | 2.5 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.3 | 2.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.3 | 3.8 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 1.0 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.3 | 1.9 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.3 | 0.9 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.3 | 4.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.3 | 3.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.3 | 1.5 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.3 | 5.1 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.3 | 2.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.3 | 1.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 2.5 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.2 | 1.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 1.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 1.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 0.9 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 2.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 0.9 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 14.1 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.2 | 1.1 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.2 | 3.3 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.2 | 1.7 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.2 | 5.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 1.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 4.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 0.8 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 3.8 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.2 | 2.0 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.2 | 43.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.2 | 7.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 1.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 0.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 0.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.5 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 1.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 3.5 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.2 | 0.6 | GO:0045938 | regulation of endocrine process(GO:0044060) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) endocrine hormone secretion(GO:0060986) |
| 0.2 | 1.1 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.2 | 0.6 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.2 | 5.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 0.9 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.2 | 0.5 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.6 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 0.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.7 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.1 | 1.4 | GO:0098869 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 1.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 2.6 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 1.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.7 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 0.7 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 1.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 2.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 0.9 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 2.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.4 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 2.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 4.1 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.1 | 0.4 | GO:0021985 | parturition(GO:0007567) neurohypophysis development(GO:0021985) maternal process involved in parturition(GO:0060137) |
| 0.1 | 4.4 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.1 | 0.5 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 4.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 3.0 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 1.8 | GO:0099590 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 4.9 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 3.7 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 0.8 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 0.4 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 1.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 1.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 5.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.1 | 0.5 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 1.9 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 2.9 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 3.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.2 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.1 | 0.4 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 1.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 1.2 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 1.6 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.1 | 2.8 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.3 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 3.0 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.1 | 2.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 16.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 0.9 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.9 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 3.1 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 0.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 1.0 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.9 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.1 | 2.1 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.1 | 0.7 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.1 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.1 | 0.5 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 1.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 4.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.1 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.1 | 1.2 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 3.6 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 1.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 3.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 2.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 2.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.9 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 1.3 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0060347 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.0 | 1.5 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.6 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 1.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 3.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.3 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.5 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 4.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.2 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.3 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 1.8 | GO:2000181 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 | 2.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 2.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.1 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.0 | 0.5 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 6.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 2.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.7 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 4.0 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 1.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 1.0 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.4 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 0.1 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.4 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.4 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 16.2 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 8.6 | GO:0005975 | carbohydrate metabolic process(GO:0005975) |
| 0.0 | 0.3 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 2.6 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 0.5 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.4 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 9.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.8 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 1.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.7 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.0 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.6 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 2.1 | 12.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.7 | 8.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 1.2 | 3.5 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.9 | 6.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.9 | 2.6 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.8 | 6.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 2.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.5 | 5.6 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.4 | 2.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.4 | 2.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.4 | 2.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.4 | 7.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.4 | 1.6 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.3 | 3.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 4.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 1.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 0.9 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.3 | 5.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.3 | 4.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.3 | 3.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 17.9 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.2 | 0.9 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 13.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 2.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 8.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 5.1 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.2 | 0.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 4.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 6.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 4.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 3.5 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 3.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 2.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.2 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 2.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 1.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 3.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 3.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 7.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 0.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 9.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 2.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 4.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 6.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 2.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.0 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 4.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 43.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 7.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 2.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 3.9 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 36.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 2.0 | GO:0060076 | postsynaptic density(GO:0014069) excitatory synapse(GO:0060076) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 14.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 3.3 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.3 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 8.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.7 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.9 | GO:0043005 | neuron projection(GO:0043005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 14.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.5 | 4.5 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 1.5 | 5.9 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 1.4 | 5.5 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 1.3 | 5.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.3 | 7.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.9 | 6.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.9 | 25.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.9 | 2.6 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.8 | 6.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.8 | 3.3 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.8 | 6.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.7 | 2.9 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.7 | 2.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.6 | 3.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.6 | 3.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.6 | 3.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.6 | 4.6 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.6 | 3.4 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.5 | 2.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.5 | 4.2 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.5 | 2.6 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.5 | 1.5 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.5 | 3.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.5 | 4.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.5 | 4.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.4 | 2.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.4 | 5.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 5.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.4 | 3.7 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.4 | 11.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 2.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.4 | 1.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 2.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 1.0 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.3 | 1.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.3 | 12.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 1.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.3 | 1.5 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.3 | 2.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 4.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 0.8 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.3 | 2.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 21.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.7 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.2 | 1.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 0.7 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 0.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 1.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 0.7 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.2 | 1.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 1.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 8.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 4.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 2.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 3.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 0.8 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.2 | 1.0 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.2 | 6.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 1.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 2.8 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.2 | 0.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 2.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.2 | 1.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 0.6 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.2 | 7.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 2.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 19.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.5 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.0 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 1.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 1.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.7 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 31.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.9 | GO:0008443 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) phosphofructokinase activity(GO:0008443) |
| 0.1 | 1.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 2.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 2.4 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.4 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.1 | 1.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 4.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 3.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 4.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.6 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 2.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.7 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.5 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 5.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 4.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.5 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 1.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 6.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 1.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.0 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.3 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 1.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.6 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.4 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 2.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 5.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.3 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 2.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.6 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.9 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.8 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.8 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 5.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 3.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 4.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 2.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.5 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.2 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 8.1 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 4.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 4.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 1.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0022839 | calcium activated cation channel activity(GO:0005227) ion gated channel activity(GO:0022839) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 1.0 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 6.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.8 | GO:0016597 | amino acid binding(GO:0016597) |
| 0.0 | 10.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 3.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.5 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 2.2 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.8 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 0.8 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.3 | 2.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 3.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 6.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 4.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 2.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 6.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 4.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 0.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 2.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 4.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 6.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.6 | 1.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.6 | 6.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 1.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 4.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.3 | 6.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.3 | 5.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.3 | 2.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 0.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 2.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 6.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 3.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 3.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 5.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 2.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 3.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 6.1 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.2 | 8.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 4.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.7 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 2.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.3 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |