Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
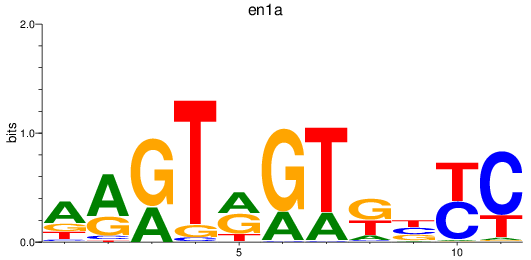
Results for en1a
Z-value: 0.75
Transcription factors associated with en1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
en1a
|
ENSDARG00000014321 | engrailed homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| en1a | dr11_v1_chr9_-_785444_785444 | 0.30 | 2.1e-01 | Click! |
Activity profile of en1a motif
Sorted Z-values of en1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_9277152 | 1.41 |
ENSDART00000189048
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr5_+_22459087 | 1.37 |
ENSDART00000134781
|
BX546499.1
|
|
| chr10_+_28428222 | 1.32 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr7_-_17297156 | 1.15 |
ENSDART00000161336
|
nitr11a
|
novel immune-type receptor 11a |
| chr14_-_41467497 | 1.01 |
ENSDART00000181220
|
mid1ip1l
|
MID1 interacting protein 1, like |
| chr7_+_55314206 | 0.91 |
ENSDART00000132852
|
ctu2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr7_+_17120811 | 0.89 |
ENSDART00000147140
|
prmt3
|
protein arginine methyltransferase 3 |
| chr17_-_6349044 | 0.85 |
ENSDART00000081707
|
oct2
|
organic cation transporter 2 |
| chr7_-_20582842 | 0.84 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr10_-_21542702 | 0.83 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr16_-_41535690 | 0.81 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr18_+_19456648 | 0.80 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr22_+_2183110 | 0.80 |
ENSDART00000159279
ENSDART00000121703 |
znf1152
|
zinc finger protein 1152 |
| chr3_+_23768898 | 0.76 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr3_-_26184018 | 0.75 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr5_-_65021736 | 0.75 |
ENSDART00000162368
ENSDART00000161876 |
anxa1c
|
annexin A1c |
| chr11_-_669558 | 0.74 |
ENSDART00000173450
|
pparg
|
peroxisome proliferator-activated receptor gamma |
| chr4_-_52165969 | 0.72 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr10_+_38512270 | 0.69 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr2_-_26590628 | 0.69 |
ENSDART00000025120
|
ndc1
|
NDC1 transmembrane nucleoporin |
| chr17_-_18797245 | 0.68 |
ENSDART00000045991
|
vrk1
|
vaccinia related kinase 1 |
| chr25_-_3836597 | 0.68 |
ENSDART00000115154
|
si:ch211-247i17.1
|
si:ch211-247i17.1 |
| chr18_-_43866526 | 0.66 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr7_+_4592867 | 0.66 |
ENSDART00000192614
|
si:dkey-83f18.15
|
si:dkey-83f18.15 |
| chr1_-_18585046 | 0.66 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr7_-_18712482 | 0.66 |
ENSDART00000173517
ENSDART00000122564 ENSDART00000173905 |
trmt10a
|
tRNA methyltransferase 10A |
| chr20_+_51833030 | 0.65 |
ENSDART00000074330
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr9_+_44994214 | 0.65 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr12_+_27096835 | 0.65 |
ENSDART00000149475
|
ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr22_+_17120473 | 0.64 |
ENSDART00000170076
ENSDART00000090217 |
frem1b
|
Fras1 related extracellular matrix 1b |
| chr12_-_10365875 | 0.64 |
ENSDART00000142386
ENSDART00000007335 |
ndc80
|
NDC80 kinetochore complex component |
| chr8_-_50888806 | 0.63 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr6_+_21684296 | 0.62 |
ENSDART00000057223
|
rhebl1
|
Ras homolog, mTORC1 binding like 1 |
| chr2_+_36606019 | 0.62 |
ENSDART00000098415
|
ing5b
|
inhibitor of growth family, member 5b |
| chr7_-_6470431 | 0.62 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr12_-_10381282 | 0.60 |
ENSDART00000152788
|
mki67
|
marker of proliferation Ki-67 |
| chr22_-_10440688 | 0.60 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr9_-_33121535 | 0.60 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr11_+_3989403 | 0.59 |
ENSDART00000082379
|
spcs1
|
signal peptidase complex subunit 1 |
| chr11_-_35756468 | 0.59 |
ENSDART00000103076
|
arl8bb
|
ADP-ribosylation factor-like 8Bb |
| chr22_+_18349794 | 0.59 |
ENSDART00000186580
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr24_+_18299175 | 0.59 |
ENSDART00000140994
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr1_+_49682312 | 0.58 |
ENSDART00000136747
|
si:ch211-149l1.2
|
si:ch211-149l1.2 |
| chr3_+_14543953 | 0.58 |
ENSDART00000161710
|
epor
|
erythropoietin receptor |
| chr9_+_21277846 | 0.58 |
ENSDART00000139620
ENSDART00000110996 ENSDART00000111899 |
lats2
|
large tumor suppressor kinase 2 |
| chr24_-_2423791 | 0.57 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr13_+_18311410 | 0.55 |
ENSDART00000036718
ENSDART00000132073 |
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr12_+_34119439 | 0.55 |
ENSDART00000032821
|
cyth1b
|
cytohesin 1b |
| chr18_+_41232719 | 0.55 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr10_-_36633882 | 0.55 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr8_-_39859688 | 0.55 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr23_+_42396934 | 0.55 |
ENSDART00000169821
|
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr23_-_16981899 | 0.54 |
ENSDART00000125472
ENSDART00000142297 ENSDART00000143180 |
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr2_-_24398324 | 0.54 |
ENSDART00000165226
|
zgc:154006
|
zgc:154006 |
| chr19_+_12649691 | 0.54 |
ENSDART00000192956
ENSDART00000088917 |
rnmt
|
RNA (guanine-7-) methyltransferase |
| chr5_-_8164439 | 0.51 |
ENSDART00000189912
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr1_-_38170997 | 0.51 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr8_+_40284973 | 0.50 |
ENSDART00000018401
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr5_-_69716501 | 0.50 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr4_-_4250317 | 0.49 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr16_+_23924040 | 0.48 |
ENSDART00000161124
|
si:dkey-7f3.14
|
si:dkey-7f3.14 |
| chr4_-_72296520 | 0.47 |
ENSDART00000182638
|
si:cabz01071911.3
|
si:cabz01071911.3 |
| chr7_+_69459759 | 0.47 |
ENSDART00000160500
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr4_-_55039830 | 0.47 |
ENSDART00000170096
|
si:dkey-56m15.3
|
si:dkey-56m15.3 |
| chr2_+_37836821 | 0.47 |
ENSDART00000143203
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr13_+_8892784 | 0.46 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr9_+_23003208 | 0.45 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr2_+_44972720 | 0.44 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr2_-_3045861 | 0.43 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr11_-_6880725 | 0.43 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr2_+_26656283 | 0.43 |
ENSDART00000133202
ENSDART00000099208 |
asph
|
aspartate beta-hydroxylase |
| chr23_+_27782071 | 0.43 |
ENSDART00000131379
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr18_-_17513426 | 0.42 |
ENSDART00000146725
|
b3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr23_-_10722664 | 0.41 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr7_-_39751540 | 0.40 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr6_+_47424266 | 0.40 |
ENSDART00000171087
|
si:ch211-286o17.1
|
si:ch211-286o17.1 |
| chr5_+_61863996 | 0.40 |
ENSDART00000082879
|
sympk
|
symplekin |
| chr2_+_22495274 | 0.40 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 VRAC subunit Da |
| chr18_-_14901437 | 0.39 |
ENSDART00000145842
ENSDART00000008035 |
trabd
|
TraB domain containing |
| chr21_+_25198637 | 0.39 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr3_+_26345732 | 0.39 |
ENSDART00000128613
|
rps15a
|
ribosomal protein S15a |
| chr22_-_37738203 | 0.39 |
ENSDART00000143190
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr23_-_25779995 | 0.39 |
ENSDART00000110670
|
si:dkey-21c19.3
|
si:dkey-21c19.3 |
| chr7_-_34062301 | 0.39 |
ENSDART00000052404
|
map2k5
|
mitogen-activated protein kinase kinase 5 |
| chr25_-_17503393 | 0.39 |
ENSDART00000061726
|
trim35-40
|
tripartite motif containing 35-40 |
| chr2_+_22531185 | 0.39 |
ENSDART00000171959
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr4_-_20108833 | 0.39 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr15_-_20233105 | 0.38 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr22_+_22303053 | 0.38 |
ENSDART00000129975
|
scamp4
|
secretory carrier membrane protein 4 |
| chr14_+_14043793 | 0.37 |
ENSDART00000164376
|
rraga
|
Ras-related GTP binding A |
| chr1_-_11913170 | 0.37 |
ENSDART00000165543
|
grk4
|
G protein-coupled receptor kinase 4 |
| chr22_+_15973122 | 0.37 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr23_-_33738945 | 0.37 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr22_-_37007365 | 0.37 |
ENSDART00000157966
|
ATP11B
|
si:dkey-211e20.10 |
| chr8_+_17168114 | 0.37 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr12_-_8070969 | 0.37 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr17_-_45734459 | 0.36 |
ENSDART00000137807
|
arf6b
|
ADP-ribosylation factor 6b |
| chr14_+_45732081 | 0.36 |
ENSDART00000110103
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr25_+_19095231 | 0.35 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr6_-_8392104 | 0.35 |
ENSDART00000081561
ENSDART00000181178 |
ilf3a
|
interleukin enhancer binding factor 3a |
| chr10_+_16165533 | 0.35 |
ENSDART00000065045
|
prrc1
|
proline-rich coiled-coil 1 |
| chr4_+_13615731 | 0.35 |
ENSDART00000141395
ENSDART00000144310 |
ccdc87
|
coiled-coil domain containing 87 |
| chr18_-_29898351 | 0.35 |
ENSDART00000138533
ENSDART00000064076 |
cmc2
|
C-x(9)-C motif containing 2 |
| chr25_+_7532627 | 0.35 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr14_-_25935167 | 0.34 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr13_-_1130096 | 0.34 |
ENSDART00000010261
|
pno1
|
partner of NOB1 homolog |
| chr4_+_25912654 | 0.34 |
ENSDART00000109508
ENSDART00000134218 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr9_-_30555725 | 0.34 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr1_-_57172294 | 0.33 |
ENSDART00000063774
|
rac1l
|
Rac family small GTPase 1, like |
| chr12_+_36416173 | 0.33 |
ENSDART00000190278
|
unk
|
unkempt family zinc finger |
| chr11_-_44898129 | 0.33 |
ENSDART00000157615
|
eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr2_-_16562505 | 0.33 |
ENSDART00000156406
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr13_+_25364324 | 0.33 |
ENSDART00000187471
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr17_-_45734231 | 0.33 |
ENSDART00000074873
|
arf6b
|
ADP-ribosylation factor 6b |
| chr6_-_54111928 | 0.32 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr19_-_32914227 | 0.32 |
ENSDART00000186115
ENSDART00000124246 |
mtdha
|
metadherin a |
| chr8_-_21054132 | 0.32 |
ENSDART00000187735
|
zgc:112962
|
zgc:112962 |
| chr23_+_36653376 | 0.31 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr16_-_45080812 | 0.31 |
ENSDART00000157377
|
si:rp71-77l1.2
|
si:rp71-77l1.2 |
| chr2_+_26498446 | 0.31 |
ENSDART00000078412
|
rps8a
|
ribosomal protein S8a |
| chr22_+_2409175 | 0.31 |
ENSDART00000141776
|
zgc:113220
|
zgc:113220 |
| chr15_-_26931541 | 0.31 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr13_-_15700060 | 0.30 |
ENSDART00000170689
ENSDART00000010986 ENSDART00000101741 ENSDART00000139124 |
ckba
|
creatine kinase, brain a |
| chr2_+_36114194 | 0.30 |
ENSDART00000113547
|
traj39
|
T-cell receptor alpha joining 39 |
| chr22_+_2533514 | 0.30 |
ENSDART00000147967
|
si:ch73-92e7.4
|
si:ch73-92e7.4 |
| chr9_+_14152211 | 0.30 |
ENSDART00000148055
|
si:ch211-67e16.11
|
si:ch211-67e16.11 |
| chr2_+_41524238 | 0.30 |
ENSDART00000122860
ENSDART00000017977 |
acvr1l
|
activin A receptor, type 1 like |
| chr20_-_48898560 | 0.30 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr21_-_31252131 | 0.30 |
ENSDART00000121946
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr1_-_17689858 | 0.29 |
ENSDART00000181078
|
cfap97
|
cilia and flagella associated protein 97 |
| chr2_+_38025260 | 0.29 |
ENSDART00000075905
|
hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr1_-_38171648 | 0.29 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr23_+_1730663 | 0.29 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr5_-_41996414 | 0.29 |
ENSDART00000143573
|
ncor1
|
nuclear receptor corepressor 1 |
| chr25_-_13023569 | 0.28 |
ENSDART00000167232
|
ccl39.1
|
chemokine (C-C motif) ligand 39, duplicate 1 |
| chr2_-_24554416 | 0.28 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr25_+_17589906 | 0.27 |
ENSDART00000167750
|
vac14
|
vac14 homolog (S. cerevisiae) |
| chr6_+_18544791 | 0.27 |
ENSDART00000167463
ENSDART00000169599 |
atad5b
|
ATPase family, AAA domain containing 5b |
| chr8_-_22531817 | 0.27 |
ENSDART00000140606
|
csde1
|
cold shock domain containing E1, RNA-binding |
| chr8_-_13211527 | 0.27 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr20_-_14718801 | 0.27 |
ENSDART00000137605
|
suco
|
SUN domain containing ossification factor |
| chr3_+_12784460 | 0.26 |
ENSDART00000168382
|
cyp2k8
|
cytochrome P450, family 2, subfamily K, polypeptide 8 |
| chr18_-_7422787 | 0.26 |
ENSDART00000081386
|
si:dkey-30c15.12
|
si:dkey-30c15.12 |
| chr9_-_25255490 | 0.26 |
ENSDART00000141502
|
htr2aa
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate a |
| chr11_-_3907937 | 0.26 |
ENSDART00000082409
|
rpn1
|
ribophorin I |
| chr5_-_55625247 | 0.26 |
ENSDART00000133156
|
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr6_+_41446541 | 0.26 |
ENSDART00000029553
ENSDART00000128756 ENSDART00000144864 |
rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr15_+_23799461 | 0.25 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr19_+_7495791 | 0.25 |
ENSDART00000010862
|
mrpl24
|
mitochondrial ribosomal protein L24 |
| chr14_-_38872536 | 0.25 |
ENSDART00000184315
ENSDART00000127479 |
gsr
|
glutathione reductase |
| chr16_+_23383316 | 0.25 |
ENSDART00000103220
|
krtcap2
|
keratinocyte associated protein 2 |
| chr14_-_21123551 | 0.24 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr12_+_17754859 | 0.24 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr11_+_24716837 | 0.24 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr5_-_13778726 | 0.24 |
ENSDART00000051655
|
snrnp27
|
small nuclear ribonucleoprotein 27 (U4/U6.U5) |
| chr22_+_36547670 | 0.24 |
ENSDART00000125991
ENSDART00000186372 |
armc9
|
armadillo repeat containing 9 |
| chr22_-_20733822 | 0.24 |
ENSDART00000133780
|
si:dkey-3k20.1
|
si:dkey-3k20.1 |
| chr5_-_28038623 | 0.24 |
ENSDART00000160013
|
zgc:113436
|
zgc:113436 |
| chr1_-_11607062 | 0.24 |
ENSDART00000158253
|
si:dkey-26i13.6
|
si:dkey-26i13.6 |
| chr4_+_25912308 | 0.23 |
ENSDART00000167845
ENSDART00000136927 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr4_-_64912026 | 0.23 |
ENSDART00000157956
|
si:ch211-234c11.2
|
si:ch211-234c11.2 |
| chr14_+_26247319 | 0.23 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr8_+_17167876 | 0.23 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr2_-_37837056 | 0.22 |
ENSDART00000158179
ENSDART00000160317 ENSDART00000171409 |
mettl17
|
methyltransferase like 17 |
| chr19_-_10810006 | 0.22 |
ENSDART00000151157
|
si:dkey-3n22.9
|
si:dkey-3n22.9 |
| chr15_-_45246911 | 0.22 |
ENSDART00000189557
ENSDART00000185291 |
CABZ01072607.1
|
|
| chr1_+_14406098 | 0.22 |
ENSDART00000030083
|
chic2
|
cysteine-rich hydrophobic domain 2 |
| chr4_-_71983381 | 0.22 |
ENSDART00000169130
|
ftr64
|
finTRIM family, member 64 |
| chr2_+_49631850 | 0.22 |
ENSDART00000114274
ENSDART00000114516 |
si:dkey-53k12.18
|
si:dkey-53k12.18 |
| chr3_-_3939785 | 0.22 |
ENSDART00000049593
|
unm_sa1506
|
un-named sa1506 |
| chr14_+_16813816 | 0.22 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr1_+_33647682 | 0.22 |
ENSDART00000114384
|
znf654
|
zinc finger protein 654 |
| chr3_+_16663373 | 0.21 |
ENSDART00000100961
|
zgc:55558
|
zgc:55558 |
| chr9_+_13733468 | 0.21 |
ENSDART00000165954
ENSDART00000160355 ENSDART00000081040 ENSDART00000138254 ENSDART00000081015 ENSDART00000141314 |
abi2a
|
abl-interactor 2a |
| chr20_-_39391833 | 0.21 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr3_-_9738985 | 0.21 |
ENSDART00000160102
|
CR339059.1
|
|
| chr3_+_3519607 | 0.21 |
ENSDART00000150922
|
cdc42ep1b
|
CDC42 effector protein (Rho GTPase binding) 1b |
| chr22_+_1285182 | 0.21 |
ENSDART00000171750
|
si:ch73-138e16.3
|
si:ch73-138e16.3 |
| chr25_+_17590095 | 0.21 |
ENSDART00000009767
|
vac14
|
vac14 homolog (S. cerevisiae) |
| chr2_-_22530969 | 0.20 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr16_-_23885482 | 0.20 |
ENSDART00000132243
|
osgin2
|
oxidative stress induced growth inhibitor family member 2 |
| chr7_-_34265481 | 0.20 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr1_-_45616242 | 0.20 |
ENSDART00000150066
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr23_+_30736895 | 0.20 |
ENSDART00000042944
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr4_+_65332578 | 0.20 |
ENSDART00000170824
|
znf1108
|
zinc finger protein 1108 |
| chr21_-_21824285 | 0.19 |
ENSDART00000030984
|
wdr73
|
WD repeat domain 73 |
| chr20_-_20312789 | 0.19 |
ENSDART00000114779
|
si:ch211-212g7.6
|
si:ch211-212g7.6 |
| chr21_-_21570134 | 0.19 |
ENSDART00000174388
|
or133-4
|
odorant receptor, family H, subfamily 133, member 4 |
| chr12_+_1385192 | 0.19 |
ENSDART00000160788
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr4_-_28958601 | 0.19 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr1_+_39597809 | 0.19 |
ENSDART00000122700
|
tenm3
|
teneurin transmembrane protein 3 |
| chr17_-_12407106 | 0.19 |
ENSDART00000183540
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr9_+_55857193 | 0.18 |
ENSDART00000160980
|
sept10
|
septin 10 |
| chr13_+_22476742 | 0.18 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr4_-_5831522 | 0.18 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr9_+_3175201 | 0.18 |
ENSDART00000179022
ENSDART00000007756 ENSDART00000182108 |
dync1i2a
|
dynein, cytoplasmic 1, intermediate chain 2a |
| chr6_+_35052721 | 0.18 |
ENSDART00000191090
ENSDART00000082940 |
uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr19_+_9091673 | 0.18 |
ENSDART00000052898
|
si:ch211-81a5.5
|
si:ch211-81a5.5 |
| chr20_-_37813863 | 0.17 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr2_+_3201345 | 0.17 |
ENSDART00000130349
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
Network of associatons between targets according to the STRING database.
First level regulatory network of en1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.2 | 0.6 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 0.6 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.2 | 0.7 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.2 | 0.9 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.7 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.8 | GO:0002825 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.4 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.1 | 0.8 | GO:0021661 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.1 | 0.5 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.5 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.4 | GO:0044003 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.1 | 0.5 | GO:1900044 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.8 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 0.4 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.1 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.3 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.9 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.6 | GO:0046620 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.0 | 0.2 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 1.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.4 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.2 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.3 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.4 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.3 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.2 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 1.2 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.7 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.3 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.4 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.8 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.3 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.4 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.2 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 0.5 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.5 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 0.5 | GO:0035032 | extrinsic component of vacuolar membrane(GO:0000306) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.4 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.3 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.0 | 0.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.0 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.2 | 0.6 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.2 | 0.7 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.4 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.4 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.9 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.5 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.2 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.9 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.6 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0047760 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0004532 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 2.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |