Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
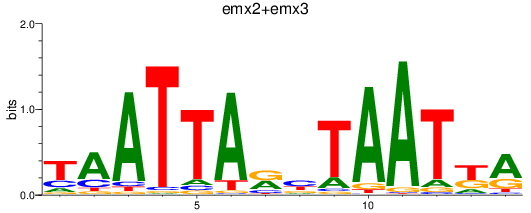
Results for emx2+emx3
Z-value: 1.01
Transcription factors associated with emx2+emx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
emx3
|
ENSDARG00000020417 | empty spiracles homeobox 3 |
|
emx2
|
ENSDARG00000039701 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| emx3 | dr11_v1_chr14_-_26377044_26377044 | -0.86 | 1.8e-06 | Click! |
| emx2 | dr11_v1_chr13_+_19322686_19322686 | -0.65 | 2.7e-03 | Click! |
Activity profile of emx2+emx3 motif
Sorted Z-values of emx2+emx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_34896956 | 2.92 |
ENSDART00000055415
|
prph2a
|
peripherin 2a (retinal degeneration, slow) |
| chr5_+_2815021 | 2.61 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr7_-_38638809 | 2.54 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr14_+_45675306 | 2.47 |
ENSDART00000105461
|
rom1b
|
retinal outer segment membrane protein 1b |
| chr7_-_38638276 | 2.30 |
ENSDART00000074463
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr25_+_14092871 | 2.27 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr17_-_12336987 | 2.01 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr6_+_39905021 | 2.00 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr25_+_31227747 | 1.93 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr25_+_34984333 | 1.73 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr10_+_45089820 | 1.73 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr23_-_306796 | 1.71 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr8_-_23780334 | 1.70 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr6_+_612594 | 1.66 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr20_-_40755614 | 1.63 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr19_+_43297546 | 1.57 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr15_+_45640906 | 1.56 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr16_-_38118003 | 1.55 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr9_-_22099536 | 1.54 |
ENSDART00000101923
|
CR391987.1
|
|
| chr3_-_32169754 | 1.50 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr11_+_6819050 | 1.49 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr4_+_25654686 | 1.48 |
ENSDART00000100714
|
acot16
|
acyl-CoA thioesterase 16 |
| chr6_-_39344259 | 1.48 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr21_+_10739846 | 1.44 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr23_-_25976656 | 1.43 |
ENSDART00000019519
|
ada
|
adenosine deaminase |
| chr5_+_37068223 | 1.33 |
ENSDART00000164279
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr15_-_14625373 | 1.31 |
ENSDART00000171841
|
slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr5_-_24238733 | 1.30 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr18_+_26829086 | 1.30 |
ENSDART00000098356
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr18_+_45483282 | 1.23 |
ENSDART00000183725
|
cngb1a
|
cyclic nucleotide gated channel beta 1a |
| chr7_+_23515966 | 1.23 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr15_+_36457888 | 1.22 |
ENSDART00000155100
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr21_-_22115136 | 1.20 |
ENSDART00000134715
ENSDART00000089246 ENSDART00000139789 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr1_-_47071979 | 1.20 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr14_+_46313396 | 1.17 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr18_-_898870 | 1.17 |
ENSDART00000151777
ENSDART00000062654 |
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr4_+_11384891 | 1.14 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr9_-_22147567 | 1.13 |
ENSDART00000110941
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr3_-_59297532 | 1.08 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr21_+_21195487 | 1.06 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr9_-_22182396 | 1.05 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr6_-_12851888 | 1.01 |
ENSDART00000056764
|
bmpr2a
|
bone morphogenetic protein receptor, type II a (serine/threonine kinase) |
| chr10_+_17776981 | 1.00 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr5_+_30624183 | 0.99 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr18_+_8833251 | 0.97 |
ENSDART00000143519
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr8_+_17009199 | 0.97 |
ENSDART00000139452
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr10_+_21522545 | 0.94 |
ENSDART00000131097
|
BX957322.1
|
|
| chr1_-_21901589 | 0.93 |
ENSDART00000140553
|
frmpd1a
|
FERM and PDZ domain containing 1a |
| chr18_+_26829362 | 0.93 |
ENSDART00000132728
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr21_-_20939488 | 0.91 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr4_-_69189894 | 0.86 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr18_-_6477662 | 0.86 |
ENSDART00000161263
|
iqsec3b
|
IQ motif and Sec7 domain 3b |
| chr7_+_15659280 | 0.85 |
ENSDART00000173414
|
mef2ab
|
myocyte enhancer factor 2ab |
| chr22_+_19277361 | 0.84 |
ENSDART00000190075
ENSDART00000141097 ENSDART00000136943 |
si:dkey-21e2.13
|
si:dkey-21e2.13 |
| chr21_-_4923427 | 0.83 |
ENSDART00000181489
|
kcnt1
|
potassium channel, subfamily T, member 1 |
| chr12_-_17712393 | 0.83 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr9_+_54039006 | 0.83 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr6_+_57541776 | 0.82 |
ENSDART00000157330
|
necab3
|
N-terminal EF-hand calcium binding protein 3 |
| chr14_-_8890437 | 0.82 |
ENSDART00000167242
|
ATG2A
|
si:ch73-45o6.2 |
| chr23_-_1660708 | 0.82 |
ENSDART00000175138
|
CU693481.1
|
|
| chr8_+_42917515 | 0.81 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr21_-_22827548 | 0.81 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr11_+_45110865 | 0.80 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr20_+_52389858 | 0.80 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr9_-_54001502 | 0.80 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr5_-_71705191 | 0.80 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr12_-_30523216 | 0.79 |
ENSDART00000152896
ENSDART00000153191 |
plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr20_-_1141722 | 0.79 |
ENSDART00000152675
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr13_+_7665890 | 0.78 |
ENSDART00000046792
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr2_+_20430366 | 0.78 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr20_+_34717403 | 0.78 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr15_-_5740358 | 0.77 |
ENSDART00000183563
|
phkg1a
|
phosphorylase kinase, gamma 1a (muscle) |
| chr18_+_39067575 | 0.77 |
ENSDART00000077724
|
gnb5b
|
guanine nucleotide binding protein (G protein), beta 5b |
| chr19_-_47323267 | 0.77 |
ENSDART00000190077
|
CU138512.1
|
|
| chr23_+_29966466 | 0.75 |
ENSDART00000143583
|
dvl1a
|
dishevelled segment polarity protein 1a |
| chr2_+_45548890 | 0.75 |
ENSDART00000113994
|
fndc7a
|
fibronectin type III domain containing 7a |
| chr6_-_30839763 | 0.74 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr6_+_39506043 | 0.73 |
ENSDART00000086260
|
CR388364.1
|
|
| chr2_-_1548330 | 0.73 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr9_+_55379283 | 0.73 |
ENSDART00000192728
|
nlgn4b
|
neuroligin 4b |
| chr2_+_42871831 | 0.73 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr25_+_3994823 | 0.72 |
ENSDART00000154020
|
eps8l2
|
EPS8 like 2 |
| chr10_+_24468922 | 0.71 |
ENSDART00000008248
ENSDART00000183510 |
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr6_-_39005133 | 0.71 |
ENSDART00000104116
ENSDART00000151750 |
vdrb
|
vitamin D receptor b |
| chr21_-_36571804 | 0.70 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr6_+_13933464 | 0.70 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr19_+_31873308 | 0.69 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr18_-_5527050 | 0.68 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr2_-_9059955 | 0.68 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr21_-_37661570 | 0.68 |
ENSDART00000174570
|
ndfip1l
|
Nedd4 family interacting protein 1, like |
| chr19_-_42045372 | 0.68 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr1_+_6862917 | 0.68 |
ENSDART00000182953
|
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr23_-_19103855 | 0.68 |
ENSDART00000016901
|
pard6b
|
par-6 partitioning defective 6 homolog beta (C. elegans) |
| chr12_+_47698356 | 0.67 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr21_-_44081540 | 0.66 |
ENSDART00000130833
|
FO704810.1
|
|
| chr20_-_5291012 | 0.65 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr9_+_29643036 | 0.65 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr9_-_22355391 | 0.64 |
ENSDART00000009115
|
crygm3
|
crystallin, gamma M3 |
| chr25_+_7671640 | 0.64 |
ENSDART00000145367
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr5_-_6377865 | 0.64 |
ENSDART00000031775
|
zgc:73226
|
zgc:73226 |
| chr10_+_33902264 | 0.64 |
ENSDART00000193793
|
fryb
|
furry homolog b (Drosophila) |
| chr9_+_2574122 | 0.64 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr25_+_19149241 | 0.64 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr16_+_9540033 | 0.63 |
ENSDART00000149574
|
ca14
|
carbonic anhydrase XIV |
| chr16_-_31622777 | 0.63 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr22_-_31060579 | 0.63 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr22_+_5176693 | 0.61 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr2_+_41926707 | 0.61 |
ENSDART00000023208
|
zgc:110183
|
zgc:110183 |
| chr25_-_25058508 | 0.61 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr18_-_48983690 | 0.60 |
ENSDART00000182359
|
FO681288.3
|
|
| chr1_-_22757145 | 0.60 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr3_-_42981739 | 0.58 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr21_+_5882300 | 0.58 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr11_+_30663300 | 0.57 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr23_-_18024543 | 0.57 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr7_-_66868543 | 0.56 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr24_-_31425799 | 0.56 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr4_+_22480169 | 0.55 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr7_+_69470442 | 0.54 |
ENSDART00000189593
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr20_-_43663494 | 0.54 |
ENSDART00000144564
|
BX470188.1
|
|
| chr1_+_44127292 | 0.54 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr3_+_39922602 | 0.54 |
ENSDART00000193937
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr11_+_42474694 | 0.54 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr7_-_74057288 | 0.53 |
ENSDART00000189740
|
CU929544.1
|
|
| chr20_+_37457642 | 0.53 |
ENSDART00000132771
|
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr25_-_27621268 | 0.53 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr20_+_32552912 | 0.53 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr21_-_4922981 | 0.53 |
ENSDART00000191819
ENSDART00000113556 ENSDART00000193269 ENSDART00000182961 |
kcnt1
|
potassium channel, subfamily T, member 1 |
| chr1_-_55785722 | 0.53 |
ENSDART00000142069
ENSDART00000043933 |
ndufb7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 |
| chr17_+_23584268 | 0.52 |
ENSDART00000192675
|
slc16a12a
|
solute carrier family 16, member 12a |
| chr11_+_40812590 | 0.52 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr12_-_6818676 | 0.52 |
ENSDART00000106391
|
pcdh15b
|
protocadherin-related 15b |
| chr1_-_9629705 | 0.52 |
ENSDART00000166601
ENSDART00000164079 |
ugt5b5
|
UDP glucuronosyltransferase 5 family, polypeptide B5 |
| chr23_+_41679586 | 0.52 |
ENSDART00000067662
|
CU914487.1
|
|
| chr24_-_33873451 | 0.51 |
ENSDART00000159840
|
asic1c
|
acid-sensing (proton-gated) ion channel 1c |
| chr1_+_18811679 | 0.51 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr22_+_19366866 | 0.50 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr3_+_7808459 | 0.50 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr1_-_9195629 | 0.50 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr22_-_14739491 | 0.50 |
ENSDART00000133385
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr25_+_31277415 | 0.50 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr15_+_34592215 | 0.50 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr7_+_13382852 | 0.50 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr6_-_39649504 | 0.49 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr1_-_17797802 | 0.48 |
ENSDART00000041215
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr15_-_46779934 | 0.48 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr7_-_50410524 | 0.48 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr22_+_5176255 | 0.48 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr11_-_45171139 | 0.47 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr10_+_21867307 | 0.47 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr17_-_26604946 | 0.47 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr2_-_4925025 | 0.46 |
ENSDART00000160257
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr9_-_27442339 | 0.46 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr1_+_10221568 | 0.45 |
ENSDART00000152424
|
npy2rl
|
neuropeptide Y receptor Y2, like |
| chr18_+_16750080 | 0.45 |
ENSDART00000136320
|
rnf141
|
ring finger protein 141 |
| chr11_-_44543082 | 0.45 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr12_+_47044707 | 0.45 |
ENSDART00000186506
|
zranb1a
|
zinc finger, RAN-binding domain containing 1a |
| chr7_+_69459759 | 0.44 |
ENSDART00000160500
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr4_+_76671012 | 0.44 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr5_-_71460556 | 0.44 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr11_-_8126223 | 0.43 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr18_+_21408794 | 0.43 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr12_-_18883079 | 0.43 |
ENSDART00000178619
|
shisa8b
|
shisa family member 8b |
| chr1_+_38858399 | 0.43 |
ENSDART00000165454
|
CU915762.1
|
|
| chr11_-_45141309 | 0.43 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr11_+_2855430 | 0.42 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr23_+_41800052 | 0.42 |
ENSDART00000141484
|
pdyn
|
prodynorphin |
| chr23_+_44741500 | 0.42 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr17_+_389218 | 0.41 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr7_-_11596450 | 0.41 |
ENSDART00000173863
|
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr23_+_11669109 | 0.41 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr24_-_26328721 | 0.41 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr11_+_3005536 | 0.41 |
ENSDART00000174539
|
cpne5b
|
copine Vb |
| chr21_-_25685739 | 0.41 |
ENSDART00000129619
ENSDART00000101205 |
phkg1b
|
phosphorylase kinase, gamma 1b (muscle) |
| chr9_+_11202552 | 0.41 |
ENSDART00000151931
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr7_+_73488035 | 0.41 |
ENSDART00000110330
|
arhgef40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr9_-_48397702 | 0.40 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr23_-_900795 | 0.40 |
ENSDART00000190517
ENSDART00000182849 ENSDART00000111456 ENSDART00000185430 |
rbm10
|
RNA binding motif protein 10 |
| chr11_-_42554290 | 0.40 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr25_+_35913614 | 0.40 |
ENSDART00000022437
|
gpia
|
glucose-6-phosphate isomerase a |
| chr2_-_10014821 | 0.40 |
ENSDART00000185525
|
CR956623.1
|
|
| chr23_-_42752550 | 0.40 |
ENSDART00000187059
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr13_+_733027 | 0.40 |
ENSDART00000149547
|
nrxn1b
|
neurexin 1b |
| chr8_+_20679759 | 0.40 |
ENSDART00000088668
|
nfic
|
nuclear factor I/C |
| chr13_-_31397987 | 0.39 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr23_+_45200481 | 0.39 |
ENSDART00000004357
ENSDART00000111126 ENSDART00000193560 ENSDART00000190476 |
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr1_-_25144439 | 0.38 |
ENSDART00000132355
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr8_+_36560019 | 0.38 |
ENSDART00000136418
ENSDART00000061378 ENSDART00000185237 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr17_-_43391499 | 0.38 |
ENSDART00000189280
|
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr3_+_40170216 | 0.38 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr5_+_35458346 | 0.38 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr21_-_22709251 | 0.38 |
ENSDART00000140032
|
si:dkeyp-69c1.9
|
si:dkeyp-69c1.9 |
| chr25_-_21816269 | 0.38 |
ENSDART00000152014
|
zgc:158222
|
zgc:158222 |
| chr21_+_18997511 | 0.37 |
ENSDART00000145591
|
rpl17
|
ribosomal protein L17 |
| chr3_-_34084387 | 0.37 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr15_+_11427620 | 0.37 |
ENSDART00000168688
|
ndufc2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 |
| chr5_+_45140914 | 0.37 |
ENSDART00000172702
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_2223837 | 0.36 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr16_-_28856112 | 0.36 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr20_-_44557037 | 0.35 |
ENSDART00000140995
|
mfsd2b
|
major facilitator superfamily domain containing 2B |
Network of associatons between targets according to the STRING database.
First level regulatory network of emx2+emx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.6 | 1.7 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.5 | 1.5 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.4 | 2.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 1.4 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) |
| 0.3 | 0.8 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.3 | 0.8 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.3 | 0.8 | GO:0019852 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.2 | 1.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 0.8 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 1.2 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 2.2 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 2.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.6 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 1.0 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 1.7 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 1.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.4 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.1 | 0.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.5 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.7 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.8 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.8 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 0.5 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.7 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 1.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.7 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.4 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.3 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 1.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.2 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.3 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.6 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 2.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 1.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 10.7 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.0 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 1.7 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 1.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 1.0 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.2 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 1.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 1.3 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.3 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.8 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 1.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.3 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.7 | GO:0060119 | inner ear receptor cell development(GO:0060119) |
| 0.0 | 0.4 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.5 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.3 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.2 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.3 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.3 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.7 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.5 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 1.0 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.0 | 0.3 | GO:0033500 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.5 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.6 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 0.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.1 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.1 | 1.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 4.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.4 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 2.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.6 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 1.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.3 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.8 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.3 | 1.7 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 2.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 1.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 1.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 1.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.7 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 1.8 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.4 | GO:0052725 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 1.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.6 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 0.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.4 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 1.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 1.1 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 1.5 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.4 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.0 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 0.2 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.8 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 4.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.3 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 2.2 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 1.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 2.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.1 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 5.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 1.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.7 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.7 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.3 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |