Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for elf3
Z-value: 0.71
Transcription factors associated with elf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
elf3
|
ENSDARG00000077982 | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| elf3 | dr11_v1_chr22_+_661711_661711 | -0.23 | 3.3e-01 | Click! |
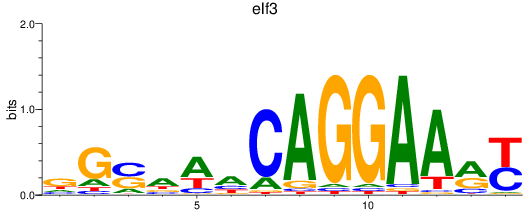
Activity profile of elf3 motif
Sorted Z-values of elf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_69482891 | 1.46 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr17_-_6730247 | 1.43 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr5_-_21524251 | 1.37 |
ENSDART00000176793
ENSDART00000040184 |
tenm1
|
teneurin transmembrane protein 1 |
| chr19_-_40192249 | 1.25 |
ENSDART00000051972
|
grn1
|
granulin 1 |
| chr6_-_10233538 | 1.18 |
ENSDART00000182004
ENSDART00000149237 ENSDART00000148876 |
xirp2a
|
xin actin binding repeat containing 2a |
| chr12_-_14551077 | 1.18 |
ENSDART00000188717
|
FO704673.1
|
|
| chr24_-_11127493 | 1.18 |
ENSDART00000144066
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr13_-_37210369 | 1.10 |
ENSDART00000140744
|
si:dkeyp-77c8.4
|
si:dkeyp-77c8.4 |
| chr24_-_38113208 | 1.08 |
ENSDART00000079003
|
crp
|
c-reactive protein, pentraxin-related |
| chr11_-_141592 | 1.04 |
ENSDART00000092787
|
cdk4
|
cyclin-dependent kinase 4 |
| chr22_-_31517300 | 0.97 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr16_+_34528409 | 0.96 |
ENSDART00000144718
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr7_+_35075847 | 0.94 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr13_+_30055171 | 0.92 |
ENSDART00000143581
ENSDART00000132027 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr6_-_17849786 | 0.92 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr16_+_14033121 | 0.92 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr14_+_26796917 | 0.90 |
ENSDART00000137695
|
klhl4
|
kelch-like family member 4 |
| chr20_-_2090789 | 0.89 |
ENSDART00000113237
|
arhgap18
|
Rho GTPase activating protein 18 |
| chr8_+_53423408 | 0.87 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr7_+_73295890 | 0.85 |
ENSDART00000174331
ENSDART00000174250 |
CABZ01083442.1
|
|
| chr10_+_3507861 | 0.84 |
ENSDART00000092684
|
rph3aa
|
rabphilin 3A homolog (mouse), a |
| chr19_+_48176745 | 0.82 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr13_+_30054996 | 0.81 |
ENSDART00000110061
ENSDART00000186045 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr1_+_2101541 | 0.81 |
ENSDART00000128187
ENSDART00000167050 ENSDART00000182153 ENSDART00000122626 ENSDART00000164488 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr3_-_5228137 | 0.80 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr10_-_22828302 | 0.77 |
ENSDART00000079459
ENSDART00000100468 |
per1a
|
period circadian clock 1a |
| chr5_+_52067723 | 0.77 |
ENSDART00000166902
|
setbp1
|
SET binding protein 1 |
| chr12_-_7824291 | 0.76 |
ENSDART00000148673
ENSDART00000149453 |
ank3b
|
ankyrin 3b |
| chr5_+_67812062 | 0.74 |
ENSDART00000158611
|
zgc:175280
|
zgc:175280 |
| chr10_-_41450367 | 0.74 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr3_+_19460991 | 0.72 |
ENSDART00000169124
|
pde6ga
|
phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog a |
| chr2_-_24068848 | 0.71 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr1_-_24349759 | 0.70 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr10_+_2568793 | 0.70 |
ENSDART00000191985
ENSDART00000081922 |
spina
|
spindlin a |
| chr23_-_1587955 | 0.68 |
ENSDART00000136037
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr1_+_157793 | 0.67 |
ENSDART00000152205
|
cul4a
|
cullin 4A |
| chr23_+_1181248 | 0.64 |
ENSDART00000170942
|
utrn
|
utrophin |
| chr9_-_22339582 | 0.63 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr1_-_47071979 | 0.61 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr4_-_9533345 | 0.61 |
ENSDART00000128501
|
si:ch73-139j3.4
|
si:ch73-139j3.4 |
| chr23_+_27068225 | 0.59 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr15_+_3808996 | 0.59 |
ENSDART00000110227
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr17_-_11418513 | 0.58 |
ENSDART00000064412
ENSDART00000151847 |
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr17_+_3993772 | 0.58 |
ENSDART00000170822
ENSDART00000168613 ENSDART00000186174 |
tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr10_+_1668106 | 0.58 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr20_-_33462961 | 0.57 |
ENSDART00000135927
|
si:dkey-65b13.1
|
si:dkey-65b13.1 |
| chr2_+_9061885 | 0.56 |
ENSDART00000028906
|
pigk
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr17_+_13664442 | 0.56 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr9_+_6079528 | 0.55 |
ENSDART00000142167
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr15_+_16525126 | 0.55 |
ENSDART00000193455
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr7_+_60161002 | 0.54 |
ENSDART00000054007
|
slc8a4b
|
solute carrier family 8 (sodium/calcium exchanger), member 4b |
| chr3_-_5228841 | 0.54 |
ENSDART00000092373
ENSDART00000182438 |
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr10_-_105100 | 0.53 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr13_+_4225173 | 0.52 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr23_+_42304602 | 0.52 |
ENSDART00000166004
|
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr7_-_6592142 | 0.52 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr22_-_16403602 | 0.52 |
ENSDART00000183417
|
lama3
|
laminin, alpha 3 |
| chr15_-_20180475 | 0.52 |
ENSDART00000048183
|
exoc3l2b
|
exocyst complex component 3-like 2b |
| chr13_-_2189761 | 0.51 |
ENSDART00000166255
|
mlip
|
muscular LMNA-interacting protein |
| chr6_-_55254786 | 0.51 |
ENSDART00000113805
|
nfatc2b
|
nuclear factor of activated T cells 2b |
| chr1_+_532766 | 0.51 |
ENSDART00000179731
ENSDART00000060944 |
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr6_+_3282809 | 0.51 |
ENSDART00000187444
ENSDART00000187407 ENSDART00000191883 |
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr8_+_50742975 | 0.50 |
ENSDART00000155664
ENSDART00000160612 |
si:ch73-6l19.2
|
si:ch73-6l19.2 |
| chr22_-_23781083 | 0.50 |
ENSDART00000166563
ENSDART00000170458 ENSDART00000166158 ENSDART00000171246 |
cfhl3
|
complement factor H like 3 |
| chr2_+_33382648 | 0.50 |
ENSDART00000137207
ENSDART00000098831 |
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr4_+_61995745 | 0.50 |
ENSDART00000171539
|
CT990567.1
|
|
| chr17_-_21993620 | 0.49 |
ENSDART00000179905
ENSDART00000078873 |
slc22a7b.3
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 3 |
| chr2_-_55779927 | 0.49 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr17_+_25290136 | 0.48 |
ENSDART00000173295
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr20_+_42761881 | 0.48 |
ENSDART00000113625
|
pimr113
|
Pim proto-oncogene, serine/threonine kinase, related 113 |
| chr3_+_46479913 | 0.48 |
ENSDART00000149755
|
tyk2
|
tyrosine kinase 2 |
| chr1_-_37383741 | 0.48 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr17_-_5610514 | 0.47 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr19_-_36234185 | 0.47 |
ENSDART00000186003
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr17_+_27134806 | 0.47 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr12_+_43585527 | 0.46 |
ENSDART00000161590
|
clrn3
|
clarin 3 |
| chr21_+_5888641 | 0.46 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr23_-_874418 | 0.46 |
ENSDART00000133906
|
rbm10
|
RNA binding motif protein 10 |
| chr5_+_42467867 | 0.46 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr7_+_65261354 | 0.45 |
ENSDART00000168388
|
bco1
|
beta-carotene oxygenase 1 |
| chr7_+_65261576 | 0.45 |
ENSDART00000169566
|
bco1
|
beta-carotene oxygenase 1 |
| chr4_-_14926637 | 0.45 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr3_+_24459709 | 0.45 |
ENSDART00000180976
|
cbx6b
|
chromobox homolog 6b |
| chr16_+_46807214 | 0.45 |
ENSDART00000185524
|
CABZ01027551.1
|
|
| chr4_+_26629368 | 0.45 |
ENSDART00000183575
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr3_+_29942338 | 0.44 |
ENSDART00000158220
|
ifi35
|
interferon-induced protein 35 |
| chr4_+_2655358 | 0.44 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr9_+_12948511 | 0.44 |
ENSDART00000135797
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr10_+_10636237 | 0.44 |
ENSDART00000136853
|
fam163b
|
family with sequence similarity 163, member B |
| chr3_-_32902138 | 0.44 |
ENSDART00000144026
ENSDART00000083874 ENSDART00000145443 ENSDART00000148239 ENSDART00000134917 |
kat7a
|
K(lysine) acetyltransferase 7a |
| chr24_+_39990695 | 0.44 |
ENSDART00000040281
|
BX323854.1
|
|
| chr20_-_51727860 | 0.43 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr10_+_21576909 | 0.43 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr24_-_1985007 | 0.43 |
ENSDART00000189870
|
PARD3 (1 of many)
|
par-3 family cell polarity regulator |
| chr19_+_1003332 | 0.43 |
ENSDART00000081779
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
| chr16_-_16590489 | 0.42 |
ENSDART00000190021
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr16_-_22251414 | 0.42 |
ENSDART00000158500
ENSDART00000179998 |
atp8b2
|
ATPase phospholipid transporting 8B2 |
| chr23_-_44233408 | 0.42 |
ENSDART00000149318
ENSDART00000085528 |
zgc:158659
|
zgc:158659 |
| chr19_-_40199081 | 0.41 |
ENSDART00000051970
ENSDART00000151079 |
grn2
|
granulin 2 |
| chr15_-_8856391 | 0.41 |
ENSDART00000008273
|
rab4b
|
RAB4B, member RAS oncogene family |
| chr1_-_55008882 | 0.41 |
ENSDART00000083572
|
zgc:136864
|
zgc:136864 |
| chr14_-_1949277 | 0.40 |
ENSDART00000159435
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr4_-_17263210 | 0.40 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr22_-_16377666 | 0.40 |
ENSDART00000161878
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr16_+_1802307 | 0.40 |
ENSDART00000180026
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr1_-_59407322 | 0.40 |
ENSDART00000161661
|
si:ch211-188p14.5
|
si:ch211-188p14.5 |
| chr11_-_43104475 | 0.40 |
ENSDART00000125368
|
acyp2
|
acylphosphatase 2, muscle type |
| chr6_-_52796212 | 0.40 |
ENSDART00000154133
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr15_+_9327252 | 0.40 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr17_-_44440832 | 0.39 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr20_-_1191910 | 0.38 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr24_-_35707552 | 0.38 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr23_-_19103855 | 0.38 |
ENSDART00000016901
|
pard6b
|
par-6 partitioning defective 6 homolog beta (C. elegans) |
| chr13_+_27316632 | 0.38 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr9_-_4598883 | 0.37 |
ENSDART00000171927
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr5_-_41124241 | 0.37 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr23_+_17981127 | 0.37 |
ENSDART00000012571
ENSDART00000145200 |
chia.6
|
chitinase, acidic.6 |
| chr20_+_2139436 | 0.37 |
ENSDART00000155311
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr4_+_4902392 | 0.37 |
ENSDART00000133866
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr6_-_55354004 | 0.37 |
ENSDART00000165911
|
pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr18_-_26894732 | 0.37 |
ENSDART00000147735
ENSDART00000188938 |
BX470164.1
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr12_-_44043285 | 0.36 |
ENSDART00000163074
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr15_-_568645 | 0.36 |
ENSDART00000156744
|
cbln18
|
cerebellin 18 |
| chr3_-_11972516 | 0.36 |
ENSDART00000140123
|
hmox2b
|
heme oxygenase 2b |
| chr3_+_57997980 | 0.36 |
ENSDART00000168477
ENSDART00000193840 |
pycr1a
|
pyrroline-5-carboxylate reductase 1a |
| chr12_+_33395748 | 0.35 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr1_-_44314 | 0.35 |
ENSDART00000160050
|
tmem39a
|
transmembrane protein 39A |
| chr4_-_37766174 | 0.35 |
ENSDART00000170066
|
si:dkey-207l24.2
|
si:dkey-207l24.2 |
| chr25_-_34979548 | 0.35 |
ENSDART00000153904
|
si:dkey-108k21.7
|
si:dkey-108k21.7 |
| chr5_-_13206878 | 0.34 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr7_-_51476276 | 0.34 |
ENSDART00000082464
|
nhsl2
|
NHS-like 2 |
| chr22_+_1586060 | 0.34 |
ENSDART00000160793
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr1_+_12301913 | 0.34 |
ENSDART00000165733
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr3_+_36366053 | 0.34 |
ENSDART00000181140
|
rgs9a
|
regulator of G protein signaling 9a |
| chr21_+_40443118 | 0.34 |
ENSDART00000190675
|
ssh2b
|
slingshot protein phosphatase 2b |
| chr19_+_42694211 | 0.33 |
ENSDART00000180153
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr2_-_32769759 | 0.33 |
ENSDART00000178951
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr14_-_11529311 | 0.33 |
ENSDART00000127208
|
si:ch211-153b23.7
|
si:ch211-153b23.7 |
| chr3_+_431208 | 0.33 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr20_+_32481348 | 0.33 |
ENSDART00000185018
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr15_-_8856785 | 0.33 |
ENSDART00000192816
|
rab4b
|
RAB4B, member RAS oncogene family |
| chr20_+_43602810 | 0.32 |
ENSDART00000142543
|
si:dkey-206f10.1
|
si:dkey-206f10.1 |
| chr14_-_2327825 | 0.32 |
ENSDART00000180328
ENSDART00000191135 ENSDART00000114302 ENSDART00000189869 |
pcdh2ab8
pcdh2aa1
|
protocadherin 2 alpha b 8 protocadherin 2 alpha a 1 |
| chr6_-_12459412 | 0.32 |
ENSDART00000090266
ENSDART00000144028 |
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr25_-_35664817 | 0.32 |
ENSDART00000148718
|
lrrk2
|
leucine-rich repeat kinase 2 |
| chr1_+_40237276 | 0.32 |
ENSDART00000037553
|
faah2a
|
fatty acid amide hydrolase 2a |
| chr22_-_3595439 | 0.32 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr11_+_38280454 | 0.32 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr23_+_19790962 | 0.32 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr20_+_42780162 | 0.32 |
ENSDART00000127069
|
pimr213
|
Pim proto-oncogene, serine/threonine kinase, related 213 |
| chr19_-_3759142 | 0.32 |
ENSDART00000170431
|
btr21
|
bloodthirsty-related gene family, member 21 |
| chr13_-_5978433 | 0.31 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr7_+_19762595 | 0.31 |
ENSDART00000130347
|
si:dkey-9k7.3
|
si:dkey-9k7.3 |
| chr3_-_30625219 | 0.31 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr22_+_26600834 | 0.30 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr12_-_33859950 | 0.30 |
ENSDART00000131181
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr18_-_42333428 | 0.30 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr21_+_25801345 | 0.30 |
ENSDART00000035062
|
nf2b
|
neurofibromin 2b (merlin) |
| chr1_+_36722122 | 0.29 |
ENSDART00000111566
|
tmem184c
|
transmembrane protein 184C |
| chr25_-_32311048 | 0.29 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr6_-_15065376 | 0.29 |
ENSDART00000087797
|
tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr23_+_17980875 | 0.29 |
ENSDART00000163452
|
chia.6
|
chitinase, acidic.6 |
| chr2_+_31492662 | 0.29 |
ENSDART00000123495
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr9_+_24125445 | 0.28 |
ENSDART00000090346
|
pgap1
|
post-GPI attachment to proteins 1 |
| chr7_+_9922607 | 0.28 |
ENSDART00000184532
ENSDART00000113396 |
cers3a
|
ceramide synthase 3a |
| chr15_-_35101143 | 0.28 |
ENSDART00000178430
ENSDART00000109593 |
bloc1s3
|
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chr8_+_8712446 | 0.27 |
ENSDART00000158674
|
elk1
|
ELK1, member of ETS oncogene family |
| chr4_-_73311831 | 0.27 |
ENSDART00000174391
|
zgc:171727
|
zgc:171727 |
| chr7_+_41748693 | 0.27 |
ENSDART00000174379
ENSDART00000052168 |
hrh3
|
histamine receptor H3 |
| chr18_-_6982499 | 0.27 |
ENSDART00000101525
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr24_+_15670013 | 0.27 |
ENSDART00000185826
|
CU929414.1
|
|
| chr21_+_9576176 | 0.27 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr3_+_52737565 | 0.26 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr3_-_34337969 | 0.26 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr9_-_54304684 | 0.26 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr2_+_25929619 | 0.26 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr11_-_21304452 | 0.25 |
ENSDART00000163008
|
RASSF5
|
si:dkey-85p17.3 |
| chr5_+_37487687 | 0.25 |
ENSDART00000184659
|
CU984579.1
|
|
| chr12_+_4573696 | 0.25 |
ENSDART00000152534
|
si:dkey-94f20.4
|
si:dkey-94f20.4 |
| chr5_-_57311037 | 0.24 |
ENSDART00000149855
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr22_+_569565 | 0.24 |
ENSDART00000037069
|
usp49
|
ubiquitin specific peptidase 49 |
| chr20_+_39250673 | 0.24 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr22_-_16758438 | 0.24 |
ENSDART00000132829
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr13_-_24794486 | 0.24 |
ENSDART00000136177
|
slka
|
STE20-like kinase a |
| chr6_+_51779863 | 0.24 |
ENSDART00000108534
|
tmem74b
|
transmembrane protein 74B |
| chr2_-_37462462 | 0.24 |
ENSDART00000145896
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr6_+_40714811 | 0.24 |
ENSDART00000153868
|
ccdc36
|
coiled-coil domain containing 36 |
| chr23_-_37835794 | 0.24 |
ENSDART00000137358
|
cd40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr6_-_442163 | 0.23 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr25_+_10947743 | 0.23 |
ENSDART00000073381
|
mhc1lfa
|
major histocompatibility complex class I LFA |
| chr15_-_2010926 | 0.23 |
ENSDART00000155688
|
dock10
|
dedicator of cytokinesis 10 |
| chr8_+_22438398 | 0.23 |
ENSDART00000135145
|
zgc:153759
|
zgc:153759 |
| chr15_-_36352983 | 0.23 |
ENSDART00000059786
|
si:dkey-23k10.5
|
si:dkey-23k10.5 |
| chr21_+_3941758 | 0.23 |
ENSDART00000181345
|
setx
|
senataxin |
| chr19_+_25476228 | 0.22 |
ENSDART00000112808
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr11_-_34166960 | 0.22 |
ENSDART00000181571
|
atp13a3
|
ATPase 13A3 |
| chr5_-_37252111 | 0.22 |
ENSDART00000185110
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr13_-_21701323 | 0.22 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr14_-_34276574 | 0.22 |
ENSDART00000021437
|
gria1a
|
glutamate receptor, ionotropic, AMPA 1a |
| chr1_+_30103297 | 0.22 |
ENSDART00000146783
|
vps8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of elf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.2 | 0.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.1 | 0.7 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.5 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.7 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 0.9 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:0045830 | response to protozoan(GO:0001562) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) defense response to protozoan(GO:0042832) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.5 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.5 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.7 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.5 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.2 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.1 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 1.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.8 | GO:0043153 | response to hydrogen peroxide(GO:0042542) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.6 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 1.0 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.9 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.2 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.0 | GO:2000108 | regulation of platelet activation(GO:0010543) regulation of platelet aggregation(GO:0090330) positive regulation of leukocyte apoptotic process(GO:2000108) |
| 0.0 | 0.2 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 1.2 | GO:0030835 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.6 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.5 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 1.0 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.3 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.3 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.7 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.9 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 1.5 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.4 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.7 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0031956 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 1.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |