Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
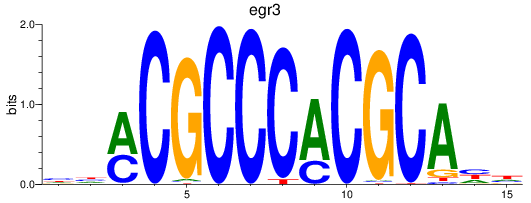
Results for egr3
Z-value: 0.33
Transcription factors associated with egr3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr3
|
ENSDARG00000089156 | early growth response 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr3 | dr11_v1_chr8_+_50727220_50727220 | 0.62 | 5.0e-03 | Click! |
Activity profile of egr3 motif
Sorted Z-values of egr3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_56422311 | 1.07 |
ENSDART00000171958
|
gpr39
|
G protein-coupled receptor 39 |
| chr25_-_207214 | 1.06 |
ENSDART00000193448
|
FP236318.3
|
|
| chr1_-_7917062 | 0.86 |
ENSDART00000177068
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr14_-_2933185 | 0.86 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr13_-_49666615 | 0.72 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr3_-_11624694 | 0.72 |
ENSDART00000127157
|
hlfa
|
hepatic leukemia factor a |
| chr6_+_2271559 | 0.71 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr13_-_31166544 | 0.69 |
ENSDART00000146250
ENSDART00000132129 ENSDART00000139591 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr9_-_33725972 | 0.68 |
ENSDART00000028225
|
mao
|
monoamine oxidase |
| chr9_-_30363770 | 0.66 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr3_-_49504023 | 0.61 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr19_-_9829965 | 0.53 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr21_+_28958471 | 0.52 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr18_+_3004572 | 0.51 |
ENSDART00000190328
|
CABZ01002690.1
|
|
| chr15_+_9294620 | 0.50 |
ENSDART00000133588
ENSDART00000140009 |
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr9_+_56422517 | 0.47 |
ENSDART00000168620
|
gpr39
|
G protein-coupled receptor 39 |
| chr19_+_5480327 | 0.46 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr2_+_38924975 | 0.45 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr14_-_7888748 | 0.43 |
ENSDART00000166293
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr5_+_6617401 | 0.41 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr5_-_46273938 | 0.40 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr7_+_67325933 | 0.39 |
ENSDART00000170575
ENSDART00000183342 |
nfat5b
|
nuclear factor of activated T cells 5b |
| chr19_-_617246 | 0.39 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr21_-_43398122 | 0.38 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr21_-_24865454 | 0.37 |
ENSDART00000142907
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr8_+_6576940 | 0.34 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr17_+_46387086 | 0.33 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr25_+_30298377 | 0.33 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr5_-_21030934 | 0.33 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr1_-_48922273 | 0.31 |
ENSDART00000150254
|
si:ch211-112b1.2
|
si:ch211-112b1.2 |
| chr23_-_29556844 | 0.29 |
ENSDART00000138021
|
rbp7a
|
retinol binding protein 7a, cellular |
| chr12_+_35650321 | 0.26 |
ENSDART00000190446
|
BX255898.1
|
|
| chr16_+_10918252 | 0.26 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr21_-_24865217 | 0.24 |
ENSDART00000101136
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr14_-_21218891 | 0.23 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr21_-_43398457 | 0.23 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr3_-_5228137 | 0.22 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr24_-_39354829 | 0.22 |
ENSDART00000169108
|
kcnj12a
|
potassium inwardly-rectifying channel, subfamily J, member 12a |
| chr17_+_48164536 | 0.22 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr18_-_19103929 | 0.21 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr22_-_16275236 | 0.20 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr10_-_15919839 | 0.20 |
ENSDART00000065032
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr10_-_26512742 | 0.20 |
ENSDART00000135951
|
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr10_-_26512993 | 0.18 |
ENSDART00000188549
ENSDART00000193316 |
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr22_+_34784075 | 0.17 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr7_-_46019756 | 0.14 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr12_-_26537145 | 0.14 |
ENSDART00000138437
ENSDART00000163931 ENSDART00000132737 |
acsf2
|
acyl-CoA synthetase family member 2 |
| chr7_-_6364168 | 0.14 |
ENSDART00000173332
|
zgc:112234
|
zgc:112234 |
| chr23_+_804119 | 0.13 |
ENSDART00000148163
|
frmd4bb
|
FERM domain containing 4Bb |
| chr24_+_3328354 | 0.13 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr12_-_44016898 | 0.13 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr25_+_1304173 | 0.12 |
ENSDART00000155229
|
rxfp3.3b
|
relaxin/insulin-like family peptide receptor 3.3b |
| chr16_-_54405976 | 0.11 |
ENSDART00000055395
|
osr2
|
odd-skipped related transciption factor 2 |
| chr20_+_27298783 | 0.11 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr13_+_12739283 | 0.10 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr6_-_53391727 | 0.10 |
ENSDART00000155483
|
si:ch211-161c3.6
|
si:ch211-161c3.6 |
| chr12_+_5215795 | 0.09 |
ENSDART00000170270
|
SLC35G1
|
si:ch211-197g18.2 |
| chr22_+_9806867 | 0.08 |
ENSDART00000137182
ENSDART00000106103 |
si:ch211-236g6.1
|
si:ch211-236g6.1 |
| chr2_+_59015878 | 0.08 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr9_-_48036 | 0.08 |
ENSDART00000165230
ENSDART00000171335 |
map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr16_+_4138331 | 0.04 |
ENSDART00000192403
ENSDART00000193036 |
mtf1
|
metal-regulatory transcription factor 1 |
| chr15_-_16121496 | 0.04 |
ENSDART00000128624
|
sgk494a
|
uncharacterized serine/threonine-protein kinase SgK494a |
| chr4_-_73587205 | 0.02 |
ENSDART00000150837
|
znf1015
|
zinc finger protein 1015 |
| chr3_+_24537023 | 0.02 |
ENSDART00000077702
|
sp100.1
|
SP110 nuclear body protein, tandem duplicate 1 |
| chr16_-_47381519 | 0.01 |
ENSDART00000032188
ENSDART00000150136 |
si:dkey-256h2.1
|
si:dkey-256h2.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 1.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.7 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.6 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.0 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0031956 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |