Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
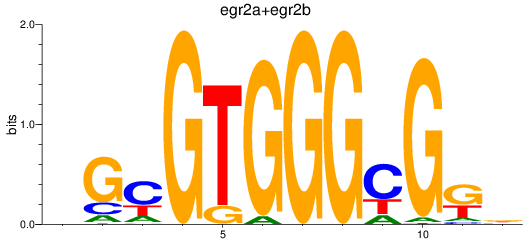
Results for egr2a+egr2b
Z-value: 1.05
Transcription factors associated with egr2a+egr2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr2b
|
ENSDARG00000042826 | early growth response 2b |
|
egr2a
|
ENSDARG00000044098 | early growth response 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr2a | dr11_v1_chr17_-_43665366_43665366 | -0.82 | 1.7e-05 | Click! |
| egr2b | dr11_v1_chr12_-_8504278_8504278 | -0.69 | 1.2e-03 | Click! |
Activity profile of egr2a+egr2b motif
Sorted Z-values of egr2a+egr2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_22535 | 6.35 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr14_+_15155684 | 5.15 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr17_+_27456804 | 3.75 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr25_+_24250247 | 3.69 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr3_-_61185746 | 3.68 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr25_-_207214 | 3.19 |
ENSDART00000193448
|
FP236318.3
|
|
| chr20_-_54462551 | 3.15 |
ENSDART00000171769
ENSDART00000169692 |
evlb
|
Enah/Vasp-like b |
| chr6_-_35446110 | 2.88 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr8_+_34731982 | 2.81 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr11_+_7324704 | 2.77 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr16_-_12173554 | 2.61 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr23_+_20422661 | 2.56 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr2_+_47582488 | 2.46 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr19_+_342094 | 2.39 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr23_-_637347 | 2.32 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr14_-_2933185 | 2.29 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr2_+_47582681 | 2.15 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr2_+_59015878 | 2.12 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr25_-_8602437 | 2.10 |
ENSDART00000171200
|
rhcgb
|
Rh family, C glycoprotein b |
| chr7_-_38612230 | 2.06 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr8_+_23165749 | 2.04 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr3_+_60007703 | 2.03 |
ENSDART00000157351
ENSDART00000153928 ENSDART00000155876 |
si:ch211-110e21.3
|
si:ch211-110e21.3 |
| chr24_-_31439841 | 2.01 |
ENSDART00000169952
|
cngb3.2
|
cyclic nucleotide gated channel beta 3, tandem duplicate 2 |
| chr14_+_2243 | 1.93 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr5_-_26093945 | 1.90 |
ENSDART00000010199
ENSDART00000145096 |
fam219ab
|
family with sequence similarity 219, member Ab |
| chr6_-_16406210 | 1.90 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr25_+_34984333 | 1.87 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr6_+_40661703 | 1.85 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr18_+_402048 | 1.84 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr6_-_15604157 | 1.81 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr24_+_3963684 | 1.79 |
ENSDART00000182959
ENSDART00000185926 ENSDART00000167043 ENSDART00000033394 |
pfkpa
|
phosphofructokinase, platelet a |
| chr17_+_46387086 | 1.78 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr8_+_24854600 | 1.75 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr21_+_28958471 | 1.75 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr16_-_12173399 | 1.72 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr6_+_42338309 | 1.72 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr21_+_5169154 | 1.68 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr12_+_18606140 | 1.67 |
ENSDART00000161128
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr11_+_77526 | 1.67 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr6_+_21740672 | 1.65 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr3_+_19299309 | 1.64 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr6_+_41503854 | 1.62 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr1_+_32528097 | 1.61 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr1_-_50438247 | 1.58 |
ENSDART00000114098
|
dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr23_+_44307996 | 1.57 |
ENSDART00000042430
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr19_-_10196370 | 1.56 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr10_+_110868 | 1.55 |
ENSDART00000135572
ENSDART00000190467 ENSDART00000056672 |
pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr24_+_26379441 | 1.53 |
ENSDART00000137786
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr9_-_54001502 | 1.49 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr20_-_14665002 | 1.47 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr13_+_12739283 | 1.47 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr1_+_1599979 | 1.47 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr11_+_1796426 | 1.46 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr7_+_67325933 | 1.43 |
ENSDART00000170575
ENSDART00000183342 |
nfat5b
|
nuclear factor of activated T cells 5b |
| chr23_-_29556844 | 1.43 |
ENSDART00000138021
|
rbp7a
|
retinol binding protein 7a, cellular |
| chr16_-_12316979 | 1.40 |
ENSDART00000182392
|
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr11_-_24191928 | 1.39 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr14_+_909769 | 1.39 |
ENSDART00000021346
ENSDART00000172777 |
arl3l2
|
ADP-ribosylation factor-like 3, like 2 |
| chr23_-_32156278 | 1.38 |
ENSDART00000157479
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr16_-_6821927 | 1.38 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr21_-_43952958 | 1.38 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr23_+_41799748 | 1.37 |
ENSDART00000144257
|
pdyn
|
prodynorphin |
| chr8_+_49975160 | 1.36 |
ENSDART00000156403
ENSDART00000080135 |
gfpt1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr5_-_43071058 | 1.34 |
ENSDART00000165546
|
si:dkey-245n4.2
|
si:dkey-245n4.2 |
| chr7_-_29534001 | 1.29 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr20_+_1398564 | 1.29 |
ENSDART00000002242
|
leg1.2
|
liver-enriched gene 1, tandem duplicate 2 |
| chr25_-_7999756 | 1.26 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr21_+_11415224 | 1.26 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr11_+_2855430 | 1.24 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr16_+_68069 | 1.22 |
ENSDART00000185385
ENSDART00000159652 |
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr20_-_45661049 | 1.21 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr3_-_28075756 | 1.20 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr23_-_12158685 | 1.20 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr20_+_13969414 | 1.20 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr7_-_13882988 | 1.19 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr17_-_20897407 | 1.18 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr2_-_31936966 | 1.18 |
ENSDART00000169484
ENSDART00000192492 ENSDART00000027689 |
amph
|
amphiphysin |
| chr16_+_52105227 | 1.16 |
ENSDART00000150025
ENSDART00000097863 |
MAN1C1
|
si:ch73-373m9.1 |
| chr2_+_30147504 | 1.13 |
ENSDART00000190947
|
kcnb2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr17_+_30587333 | 1.12 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr1_-_44940830 | 1.12 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr7_+_38510197 | 1.11 |
ENSDART00000173468
ENSDART00000100479 |
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr1_-_7917062 | 1.11 |
ENSDART00000177068
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr6_-_15604417 | 1.11 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr10_-_15053507 | 1.10 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr21_-_43949208 | 1.10 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr19_+_14573998 | 1.10 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr2_+_38924975 | 1.09 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr11_+_30161168 | 1.09 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr4_+_19534833 | 1.08 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr20_-_53366137 | 1.08 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr24_+_32472155 | 1.07 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr7_-_52849913 | 1.06 |
ENSDART00000174133
ENSDART00000172951 |
map1aa
|
microtubule-associated protein 1Aa |
| chr3_+_16762483 | 1.06 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr16_+_10918252 | 1.05 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr5_-_68022631 | 1.04 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr4_+_2619132 | 1.04 |
ENSDART00000128807
|
gpr22a
|
G protein-coupled receptor 22a |
| chr13_-_37653840 | 1.04 |
ENSDART00000143806
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr2_-_155270 | 1.03 |
ENSDART00000131177
|
adcy1b
|
adenylate cyclase 1b |
| chr16_-_2414063 | 1.02 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr10_+_1668106 | 1.02 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr11_+_3254524 | 1.01 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr6_-_25181012 | 1.00 |
ENSDART00000161585
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr23_+_43834376 | 0.99 |
ENSDART00000148989
|
si:ch211-149b19.4
|
si:ch211-149b19.4 |
| chr7_-_7845540 | 0.99 |
ENSDART00000166280
|
cxcl8b.1
|
chemokine (C-X-C motif) ligand 8b, duplicate 1 |
| chr3_+_16722014 | 0.99 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr18_+_41495841 | 0.99 |
ENSDART00000098671
|
si:ch211-203b8.6
|
si:ch211-203b8.6 |
| chr1_-_46632948 | 0.99 |
ENSDART00000148893
ENSDART00000053232 |
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr25_+_37348730 | 0.98 |
ENSDART00000156639
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr8_-_22739757 | 0.97 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr19_+_5480327 | 0.97 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr24_-_17892325 | 0.97 |
ENSDART00000154039
ENSDART00000185619 ENSDART00000178326 |
cntnap2a
|
contactin associated protein like 2a |
| chr18_-_16179129 | 0.96 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr15_+_9294620 | 0.95 |
ENSDART00000133588
ENSDART00000140009 |
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr15_-_47193564 | 0.94 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr7_-_6458222 | 0.94 |
ENSDART00000172861
|
zgc:165555
|
zgc:165555 |
| chr15_-_5467477 | 0.94 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr19_+_9305964 | 0.93 |
ENSDART00000136241
|
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr3_+_22273123 | 0.93 |
ENSDART00000044157
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr21_-_43398122 | 0.93 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr24_+_35564668 | 0.92 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr12_+_2522642 | 0.92 |
ENSDART00000152567
|
frmpd2
|
FERM and PDZ domain containing 2 |
| chr25_-_19395476 | 0.91 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr13_-_49666615 | 0.91 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr11_-_33919309 | 0.91 |
ENSDART00000078202
|
phka2
|
phosphorylase kinase, alpha 2 (liver) |
| chr3_+_40315410 | 0.91 |
ENSDART00000083241
ENSDART00000132827 |
slc29a4
|
solute carrier family 29 (equilibrative nucleoside transporter), member 4 |
| chr14_-_21218891 | 0.91 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr16_+_10963602 | 0.91 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr2_-_48966431 | 0.91 |
ENSDART00000147948
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr3_+_51684963 | 0.90 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr21_+_7900107 | 0.90 |
ENSDART00000056560
|
ch25hl2
|
cholesterol 25-hydroxylase like 2 |
| chr23_+_46183410 | 0.90 |
ENSDART00000167596
ENSDART00000151149 ENSDART00000150896 |
btr31
|
bloodthirsty-related gene family, member 31 |
| chr1_-_59176949 | 0.89 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr25_-_10564721 | 0.89 |
ENSDART00000154776
|
galn
|
galanin/GMAP prepropeptide |
| chr24_+_2519761 | 0.89 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr5_+_51443009 | 0.88 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr1_-_22861348 | 0.87 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr7_-_10560964 | 0.87 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr5_-_1999417 | 0.87 |
ENSDART00000155437
ENSDART00000145781 |
si:ch211-160e1.5
|
si:ch211-160e1.5 |
| chr16_+_17389116 | 0.87 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr15_+_3284416 | 0.86 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr25_-_35046114 | 0.85 |
ENSDART00000185267
|
zgc:165555
|
zgc:165555 |
| chr12_-_15620090 | 0.85 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr11_+_6819050 | 0.84 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr3_-_30061985 | 0.84 |
ENSDART00000189583
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr15_-_16121496 | 0.84 |
ENSDART00000128624
|
sgk494a
|
uncharacterized serine/threonine-protein kinase SgK494a |
| chr17_-_20897250 | 0.83 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr23_+_41800052 | 0.82 |
ENSDART00000141484
|
pdyn
|
prodynorphin |
| chr1_-_19845378 | 0.81 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr5_+_31820264 | 0.81 |
ENSDART00000110974
|
zdhhc12b
|
zinc finger, DHHC-type containing 12b |
| chr7_+_30867008 | 0.81 |
ENSDART00000193106
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr13_+_31757331 | 0.81 |
ENSDART00000044282
|
hif1aa
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) a |
| chr8_-_14144707 | 0.81 |
ENSDART00000148061
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr13_+_51853716 | 0.81 |
ENSDART00000175341
ENSDART00000187855 |
LT631684.1
|
|
| chr14_+_11430796 | 0.80 |
ENSDART00000165275
|
si:ch211-153b23.3
|
si:ch211-153b23.3 |
| chr13_-_40238813 | 0.80 |
ENSDART00000044963
|
loxl4
|
lysyl oxidase-like 4 |
| chr21_-_43398457 | 0.78 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr22_+_883678 | 0.78 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr10_+_32646402 | 0.78 |
ENSDART00000137244
|
zbtb21
|
zinc finger and BTB domain containing 21 |
| chr7_-_6352569 | 0.78 |
ENSDART00000173074
|
zgc:165555
|
zgc:165555 |
| chr23_+_16633951 | 0.77 |
ENSDART00000109537
ENSDART00000193323 |
snphb
|
syntaphilin b |
| chr1_+_31942961 | 0.77 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr11_-_33919014 | 0.77 |
ENSDART00000181203
|
phka2
|
phosphorylase kinase, alpha 2 (liver) |
| chr23_-_32100106 | 0.77 |
ENSDART00000044658
|
letmd1
|
LETM1 domain containing 1 |
| chr18_+_15106518 | 0.77 |
ENSDART00000168639
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr21_-_7882905 | 0.76 |
ENSDART00000056561
|
s100z
|
S100 calcium binding protein Z |
| chr10_+_252425 | 0.76 |
ENSDART00000059478
|
lrrc32
|
leucine rich repeat containing 32 |
| chr11_-_37880492 | 0.75 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr15_+_47746176 | 0.75 |
ENSDART00000154481
ENSDART00000160914 |
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr10_+_2234283 | 0.75 |
ENSDART00000136363
|
cntnap3
|
contactin associated protein like 3 |
| chr10_+_26571174 | 0.75 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr15_+_46344655 | 0.75 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr9_+_51784922 | 0.73 |
ENSDART00000125850
|
cd302
|
CD302 molecule |
| chr11_-_33960318 | 0.73 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr19_-_36675023 | 0.73 |
ENSDART00000132471
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr11_+_3254252 | 0.73 |
ENSDART00000123568
|
pmela
|
premelanosome protein a |
| chr25_+_34888886 | 0.72 |
ENSDART00000035245
|
spire2
|
spire-type actin nucleation factor 2 |
| chr7_-_20241346 | 0.72 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr12_-_13886952 | 0.72 |
ENSDART00000110503
|
adam11
|
ADAM metallopeptidase domain 11 |
| chr10_-_19497914 | 0.71 |
ENSDART00000132084
|
si:ch211-127i16.2
|
si:ch211-127i16.2 |
| chr3_-_49504023 | 0.70 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr2_+_37480669 | 0.70 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr18_+_12147971 | 0.70 |
ENSDART00000162067
ENSDART00000168386 |
fgd4a
|
FYVE, RhoGEF and PH domain containing 4a |
| chr6_-_8580857 | 0.70 |
ENSDART00000138858
ENSDART00000041142 |
myh11a
|
myosin, heavy chain 11a, smooth muscle |
| chr2_-_24603325 | 0.70 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr8_-_30338872 | 0.69 |
ENSDART00000137583
|
dock8
|
dedicator of cytokinesis 8 |
| chr19_-_6193448 | 0.69 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr1_-_28629471 | 0.69 |
ENSDART00000121758
|
ednrba
|
endothelin receptor Ba |
| chr17_-_14726824 | 0.69 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr22_+_18786797 | 0.68 |
ENSDART00000141864
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr4_-_16644708 | 0.68 |
ENSDART00000042307
|
sinhcaf
|
SIN3-HDAC complex associated factor |
| chr24_-_14711597 | 0.68 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr3_+_16976095 | 0.68 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr18_-_21806613 | 0.67 |
ENSDART00000145721
|
nrn1la
|
neuritin 1-like a |
| chr25_+_1304173 | 0.67 |
ENSDART00000155229
|
rxfp3.3b
|
relaxin/insulin-like family peptide receptor 3.3b |
| chr25_-_10565006 | 0.66 |
ENSDART00000130608
ENSDART00000190212 |
galn
|
galanin/GMAP prepropeptide |
| chr9_-_2573121 | 0.65 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr2_-_7431590 | 0.65 |
ENSDART00000185699
|
asip2b
|
agouti signaling protein, nonagouti homolog (mouse) 2b |
| chr12_-_41684729 | 0.64 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr8_+_1284784 | 0.63 |
ENSDART00000061663
|
fbxl17
|
F-box and leucine-rich repeat protein 17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr2a+egr2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.6 | 1.8 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.6 | 1.7 | GO:0097435 | fibril organization(GO:0097435) |
| 0.6 | 2.8 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.5 | 1.6 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.5 | 4.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.5 | 2.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 2.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.4 | 1.6 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.4 | 1.4 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 1.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 1.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 | 1.7 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.3 | 0.8 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.3 | 1.6 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.3 | 1.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.2 | 0.9 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.2 | 1.9 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 0.6 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.2 | 3.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 3.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 1.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 1.0 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.2 | 1.4 | GO:0071632 | optomotor response(GO:0071632) |
| 0.2 | 1.9 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 1.2 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 1.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 | 0.8 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 1.4 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 1.9 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 2.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.7 | GO:0071333 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 0.7 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 3.8 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.1 | 2.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 2.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.9 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 3.2 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.1 | 0.6 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.3 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 4.2 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 0.9 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 2.5 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.7 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.1 | 2.6 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 0.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.7 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 1.8 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.8 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.7 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.1 | 1.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.8 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.1 | 0.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.9 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.3 | GO:0019860 | uracil catabolic process(GO:0006212) beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) uracil metabolic process(GO:0019860) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.2 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 1.7 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 1.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.0 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.4 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.1 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.0 | 0.6 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 1.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.8 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 2.7 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.8 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 3.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.1 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.9 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.7 | GO:0050935 | iridophore differentiation(GO:0050935) |
| 0.0 | 1.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.5 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 2.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.3 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.0 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0097037 | heme export(GO:0097037) |
| 0.0 | 0.4 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.5 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.6 | GO:0021761 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 1.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.1 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 1.4 | GO:0050769 | positive regulation of neurogenesis(GO:0050769) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.7 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 1.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 1.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 1.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 3.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.7 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 1.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 2.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 5.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 1.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.9 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 2.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 4.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 6.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 4.2 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0098978 | glutamatergic synapse(GO:0098978) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.6 | 4.7 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.4 | 1.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 2.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.4 | 1.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.4 | 1.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 3.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 1.8 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 1.4 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.3 | 0.8 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 1.0 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.2 | 1.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 2.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 0.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 1.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 1.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 0.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 2.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.2 | 1.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 2.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 1.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.8 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 2.0 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 0.5 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 0.6 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.6 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.9 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 1.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.9 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 0.4 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 2.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 2.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 2.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.3 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 1.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 3.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.9 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 1.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 1.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.6 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.0 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 2.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.5 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 2.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 2.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 3.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 1.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 1.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 2.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |