Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
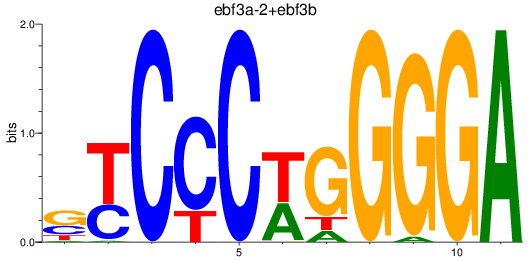
Results for ebf3a-2+ebf3b
Z-value: 0.68
Transcription factors associated with ebf3a-2+ebf3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ebf3b
|
ENSDARG00000091287 | EBF transcription factor 3b |
|
ebf3a-2
|
ENSDARG00000100244 | EBF transcription factor 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ebf3b | dr11_v1_chr17_-_20228610_20228610 | 0.64 | 3.0e-03 | Click! |
| ebf3a | dr11_v1_chr12_+_42574148_42574148 | 0.20 | 4.1e-01 | Click! |
Activity profile of ebf3a-2+ebf3b motif
Sorted Z-values of ebf3a-2+ebf3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_21471022 | 1.37 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr13_-_16226312 | 0.87 |
ENSDART00000163952
|
zgc:110045
|
zgc:110045 |
| chr23_+_16948049 | 0.86 |
ENSDART00000139449
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr17_-_22324727 | 0.80 |
ENSDART00000160341
|
CU104709.1
|
|
| chr7_+_7630409 | 0.77 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr6_+_54711306 | 0.74 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr21_-_37194669 | 0.73 |
ENSDART00000192748
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr11_+_41981959 | 0.71 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr3_-_55328548 | 0.71 |
ENSDART00000082944
|
dock6
|
dedicator of cytokinesis 6 |
| chr15_+_59417 | 0.69 |
ENSDART00000187260
|
AXL
|
AXL receptor tyrosine kinase |
| chr17_+_17955063 | 0.66 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr21_-_24632778 | 0.61 |
ENSDART00000132533
ENSDART00000058370 |
arhgap32b
|
Rho GTPase activating protein 32b |
| chr19_-_336777 | 0.54 |
ENSDART00000138837
|
golph3l
|
golgi phosphoprotein 3-like |
| chr19_+_19786117 | 0.53 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr3_-_25813426 | 0.52 |
ENSDART00000039482
|
ntn1b
|
netrin 1b |
| chr3_+_19248973 | 0.51 |
ENSDART00000174668
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr16_-_9869056 | 0.51 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr15_+_1372343 | 0.50 |
ENSDART00000152285
|
schip1
|
schwannomin interacting protein 1 |
| chr23_+_21455152 | 0.50 |
ENSDART00000158511
ENSDART00000161321 ENSDART00000160731 ENSDART00000137573 |
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr20_+_25711718 | 0.49 |
ENSDART00000033436
|
cep135
|
centrosomal protein 135 |
| chr23_+_2704793 | 0.47 |
ENSDART00000147953
|
ncoa6
|
nuclear receptor coactivator 6 |
| chr8_+_26293673 | 0.46 |
ENSDART00000144977
|
mgll
|
monoglyceride lipase |
| chr3_-_55649319 | 0.46 |
ENSDART00000176127
|
axin2
|
axin 2 (conductin, axil) |
| chr25_+_35212919 | 0.45 |
ENSDART00000180127
|
ano5a
|
anoctamin 5a |
| chr21_-_37194839 | 0.45 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr16_-_47245563 | 0.45 |
ENSDART00000190225
|
LO017877.2
|
|
| chr21_-_37194365 | 0.45 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr5_+_38900249 | 0.44 |
ENSDART00000097856
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr7_-_52558495 | 0.44 |
ENSDART00000138263
ENSDART00000009938 ENSDART00000174292 ENSDART00000174218 ENSDART00000174335 |
tcf12
|
transcription factor 12 |
| chr8_-_21988833 | 0.44 |
ENSDART00000167708
|
nphp4
|
nephronophthisis 4 |
| chr19_+_9459050 | 0.42 |
ENSDART00000186419
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr16_+_20198064 | 0.42 |
ENSDART00000006037
|
colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr7_-_18554603 | 0.42 |
ENSDART00000108938
|
msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr22_-_23545307 | 0.41 |
ENSDART00000166915
|
crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr5_+_57726425 | 0.41 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr3_-_43646733 | 0.40 |
ENSDART00000180959
|
axin1
|
axin 1 |
| chr17_+_38566717 | 0.40 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr6_+_58406014 | 0.40 |
ENSDART00000044241
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr10_-_27199135 | 0.40 |
ENSDART00000189511
ENSDART00000180314 |
auts2a
|
autism susceptibility candidate 2a |
| chr23_+_21459263 | 0.40 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr12_+_31673588 | 0.38 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr3_-_28665291 | 0.38 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr6_+_7250824 | 0.37 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr18_+_6054816 | 0.37 |
ENSDART00000113668
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr2_-_39675829 | 0.36 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr15_-_2734560 | 0.36 |
ENSDART00000153853
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr13_-_40316367 | 0.35 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr18_+_13248956 | 0.34 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr5_+_23634665 | 0.34 |
ENSDART00000125161
|
cx39.9
|
connexin 39.9 |
| chr19_+_10979983 | 0.34 |
ENSDART00000142975
|
si:ch1073-70f20.1
|
si:ch1073-70f20.1 |
| chr16_-_34212912 | 0.33 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr17_-_21441464 | 0.32 |
ENSDART00000031490
|
vax1
|
ventral anterior homeobox 1 |
| chr18_+_9382847 | 0.32 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr1_+_15062 | 0.31 |
ENSDART00000169005
|
cep97
|
centrosomal protein 97 |
| chr6_+_2951645 | 0.31 |
ENSDART00000183181
ENSDART00000181271 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr4_-_9008846 | 0.31 |
ENSDART00000164632
ENSDART00000091876 |
scube1
|
signal peptide, CUB domain, EGF-like 1 |
| chr7_-_52334840 | 0.31 |
ENSDART00000174173
|
CR938716.1
|
|
| chr9_-_33063083 | 0.30 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr18_+_30441740 | 0.30 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
| chr21_-_43665537 | 0.30 |
ENSDART00000157610
|
si:dkey-229d11.3
|
si:dkey-229d11.3 |
| chr16_-_25380903 | 0.30 |
ENSDART00000086375
ENSDART00000188587 |
adnp2a
|
ADNP homeobox 2a |
| chr4_+_8638622 | 0.30 |
ENSDART00000186829
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr25_+_4541211 | 0.30 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr13_+_29193649 | 0.30 |
ENSDART00000045951
|
unc5b
|
unc-5 netrin receptor B |
| chr11_+_7580079 | 0.30 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr17_-_30521043 | 0.29 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr23_+_20931030 | 0.28 |
ENSDART00000167014
|
pax7b
|
paired box 7b |
| chr20_+_38201644 | 0.28 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr25_+_34915762 | 0.26 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr22_+_2844865 | 0.25 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr19_+_19756425 | 0.25 |
ENSDART00000167606
|
hoxa3a
|
homeobox A3a |
| chr17_-_33407843 | 0.25 |
ENSDART00000129416
|
BX323819.1
|
|
| chr11_-_25324534 | 0.25 |
ENSDART00000158598
|
si:ch211-232b12.5
|
si:ch211-232b12.5 |
| chr21_-_27213166 | 0.25 |
ENSDART00000146959
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr10_-_42237304 | 0.24 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr1_-_35924495 | 0.24 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr3_-_3496738 | 0.23 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr10_+_31809226 | 0.22 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr9_-_33081781 | 0.22 |
ENSDART00000165748
|
zgc:172053
|
zgc:172053 |
| chr2_+_37110504 | 0.22 |
ENSDART00000132794
ENSDART00000042974 |
slc1a8b
|
solute carrier family 1 (glutamate transporter), member 8b |
| chr7_+_38811800 | 0.22 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr10_+_10972795 | 0.21 |
ENSDART00000127331
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr17_+_53425730 | 0.21 |
ENSDART00000127617
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr8_+_30671060 | 0.21 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr2_-_32457919 | 0.21 |
ENSDART00000132792
ENSDART00000041319 |
slc4a2a
|
solute carrier family 4 (anion exchanger), member 2a |
| chr2_-_10563576 | 0.20 |
ENSDART00000185818
ENSDART00000190887 |
ccdc18
|
coiled-coil domain containing 18 |
| chr15_-_23936276 | 0.19 |
ENSDART00000137569
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr25_+_34915576 | 0.19 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr5_-_62801879 | 0.18 |
ENSDART00000191749
|
adora2b
|
adenosine A2b receptor |
| chr24_-_23942722 | 0.18 |
ENSDART00000080810
|
arxa
|
aristaless related homeobox a |
| chr8_+_21297217 | 0.18 |
ENSDART00000142836
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr25_+_31323978 | 0.18 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr24_+_4977862 | 0.17 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr20_-_53321499 | 0.17 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr24_+_4978055 | 0.17 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr4_-_38259998 | 0.17 |
ENSDART00000172389
|
znf1093
|
zinc finger protein 1093 |
| chr23_-_16692312 | 0.17 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr1_+_40562540 | 0.16 |
ENSDART00000122059
|
scoca
|
short coiled-coil protein a |
| chr8_-_15182232 | 0.16 |
ENSDART00000138855
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr2_-_10943093 | 0.16 |
ENSDART00000148999
|
ssbp3a
|
single stranded DNA binding protein 3a |
| chr23_-_5032587 | 0.16 |
ENSDART00000163903
|
kcna2b
|
potassium voltage-gated channel, shaker-related subfamily, member 2b |
| chr20_+_46371458 | 0.16 |
ENSDART00000152912
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr8_-_1051438 | 0.15 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr18_+_40471826 | 0.15 |
ENSDART00000098806
|
ugt5c3
|
UDP glucuronosyltransferase 5 family, polypeptide C3 |
| chr17_+_25187670 | 0.15 |
ENSDART00000190873
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr9_-_14137295 | 0.15 |
ENSDART00000127640
ENSDART00000189018 ENSDART00000188985 |
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr8_+_8196087 | 0.14 |
ENSDART00000026965
|
plxnb3
|
plexin B3 |
| chr8_+_48858132 | 0.14 |
ENSDART00000124737
ENSDART00000079644 |
tp73
|
tumor protein p73 |
| chr24_+_6353394 | 0.14 |
ENSDART00000165118
|
CR352329.1
|
|
| chr24_+_27023616 | 0.14 |
ENSDART00000089541
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| chr13_+_17694845 | 0.13 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr16_-_30927764 | 0.13 |
ENSDART00000156804
|
slc45a4
|
solute carrier family 45, member 4 |
| chr2_-_144393 | 0.13 |
ENSDART00000156008
|
adcy1b
|
adenylate cyclase 1b |
| chr10_+_11286741 | 0.12 |
ENSDART00000187822
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr6_+_102506 | 0.12 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr14_-_36397768 | 0.11 |
ENSDART00000185199
ENSDART00000052562 |
spata4
|
spermatogenesis associated 4 |
| chr15_+_35718354 | 0.11 |
ENSDART00000110003
|
nyap2a
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2a |
| chr22_-_23253481 | 0.11 |
ENSDART00000054807
|
lhx9
|
LIM homeobox 9 |
| chr13_+_33368140 | 0.11 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr5_-_62801724 | 0.11 |
ENSDART00000144469
|
adora2b
|
adenosine A2b receptor |
| chr3_-_10621391 | 0.11 |
ENSDART00000058834
|
map2k4a
|
mitogen-activated protein kinase kinase 4a |
| chr9_-_31278048 | 0.10 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr7_-_40630698 | 0.10 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr23_-_36003282 | 0.10 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr17_+_49500820 | 0.10 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr17_-_29736019 | 0.10 |
ENSDART00000183483
|
ush2a
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr18_-_47103131 | 0.10 |
ENSDART00000188553
|
CABZ01039304.1
|
|
| chr20_+_2039518 | 0.10 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr4_-_9579299 | 0.10 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr22_+_11775269 | 0.09 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr17_+_24222190 | 0.09 |
ENSDART00000181698
ENSDART00000189411 |
ehbp1
|
EH domain binding protein 1 |
| chr11_-_3308569 | 0.09 |
ENSDART00000036581
|
cdk2
|
cyclin-dependent kinase 2 |
| chr6_+_55277419 | 0.08 |
ENSDART00000083670
|
CABZ01041604.1
|
|
| chr8_+_29856013 | 0.08 |
ENSDART00000061981
ENSDART00000149610 |
hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr18_-_7112882 | 0.08 |
ENSDART00000176292
|
si:dkey-88e18.8
|
si:dkey-88e18.8 |
| chr23_+_18527028 | 0.07 |
ENSDART00000190263
ENSDART00000104545 |
avpr2ab
|
arginine vasopressin receptor 2a, duplicate b |
| chr12_+_34763795 | 0.07 |
ENSDART00000153097
|
slc38a10
|
solute carrier family 38, member 10 |
| chr3_+_23247325 | 0.07 |
ENSDART00000114190
|
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr17_+_45454943 | 0.07 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr10_+_11286925 | 0.07 |
ENSDART00000143615
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr22_+_11756040 | 0.07 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr25_+_9003879 | 0.06 |
ENSDART00000186228
|
rag1
|
recombination activating gene 1 |
| chr11_-_27501027 | 0.06 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr9_-_98982 | 0.06 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr6_-_8399486 | 0.06 |
ENSDART00000189496
|
ptger1a
|
prostaglandin E receptor 1a (subtype EP1) |
| chr4_-_760560 | 0.06 |
ENSDART00000103601
|
agbl5
|
ATP/GTP binding protein-like 5 |
| chr7_+_54617060 | 0.05 |
ENSDART00000158898
|
fgf4
|
fibroblast growth factor 4 |
| chr10_-_7636340 | 0.05 |
ENSDART00000166234
|
ubxn8
|
UBX domain protein 8 |
| chr1_+_36911471 | 0.05 |
ENSDART00000148640
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr20_+_6590220 | 0.05 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr7_+_39410393 | 0.03 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr9_+_32930622 | 0.03 |
ENSDART00000100928
|
cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr12_+_18906407 | 0.03 |
ENSDART00000105854
|
josd1
|
Josephin domain containing 1 |
| chr13_+_502230 | 0.03 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr7_+_39410180 | 0.03 |
ENSDART00000168641
|
CT030188.1
|
|
| chr8_-_33154677 | 0.03 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr5_-_25174420 | 0.03 |
ENSDART00000141554
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr21_-_40557281 | 0.02 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr13_+_17702853 | 0.02 |
ENSDART00000145438
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr4_+_5193198 | 0.02 |
ENSDART00000067388
|
fgf23
|
fibroblast growth factor 23 |
| chr9_-_37637179 | 0.02 |
ENSDART00000188143
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr25_+_9003230 | 0.02 |
ENSDART00000180330
ENSDART00000142917 |
rag1
|
recombination activating gene 1 |
| chr13_-_4018888 | 0.02 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr9_-_7673856 | 0.02 |
ENSDART00000102715
|
tuba8l3
|
tubulin, alpha 8 like 3 |
| chr7_-_33949246 | 0.01 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr16_-_26125304 | 0.01 |
ENSDART00000176517
|
lipeb
|
lipase, hormone-sensitive b |
| chr16_+_32014552 | 0.01 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr5_+_27267186 | 0.01 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr13_-_31622195 | 0.01 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr7_-_38687431 | 0.01 |
ENSDART00000074490
|
aplnr2
|
apelin receptor 2 |
| chr11_-_22303678 | 0.00 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr16_-_36093776 | 0.00 |
ENSDART00000159134
|
OSCP1
|
organic solute carrier partner 1 |
| chr16_+_38138246 | 0.00 |
ENSDART00000131694
|
selenbp1
|
selenium binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ebf3a-2+ebf3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.2 | 0.5 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 2.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.7 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.4 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.5 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.5 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.3 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.7 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.3 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.3 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.3 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.3 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.3 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.0 | GO:0097377 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.7 | GO:0048821 | erythrocyte development(GO:0048821) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 1.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |