Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
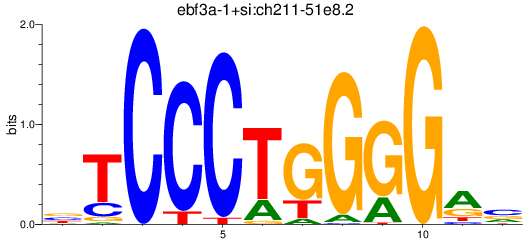
Results for ebf3a-1+si:ch211-51e8.2
Z-value: 0.84
Transcription factors associated with ebf3a-1+si:ch211-51e8.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_ch211-51e8.2
|
ENSDARG00000092635 | si_ch211-51e8.2 |
|
ebf3a-1
|
ENSDARG00000099849 | EBF transcription factor 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ebf3a | dr11_v1_chr14_+_35013656_35013656 | 0.72 | 4.5e-04 | Click! |
| EBF1 (1 of many) | dr11_v1_chr21_-_33995213_33995213 | 0.61 | 5.1e-03 | Click! |
Activity profile of ebf3a-1+si:ch211-51e8.2 motif
Sorted Z-values of ebf3a-1+si:ch211-51e8.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_9318891 | 3.18 |
ENSDART00000137364
|
si:dkey-33c12.3
|
si:dkey-33c12.3 |
| chr10_-_25769334 | 2.78 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| chr5_-_38161033 | 2.58 |
ENSDART00000145907
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr16_-_36834505 | 2.55 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr12_-_35787801 | 2.40 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr23_+_40460333 | 2.38 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr14_+_25817246 | 2.34 |
ENSDART00000136733
|
glra1
|
glycine receptor, alpha 1 |
| chr12_-_26406323 | 2.20 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr19_-_26863626 | 2.12 |
ENSDART00000145568
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr10_-_27049170 | 2.00 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr23_-_16692312 | 1.92 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr2_-_37465517 | 1.82 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr2_+_5371492 | 1.74 |
ENSDART00000139762
|
si:ch1073-184j22.1
|
si:ch1073-184j22.1 |
| chr13_-_16226312 | 1.73 |
ENSDART00000163952
|
zgc:110045
|
zgc:110045 |
| chr4_+_11375894 | 1.69 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr12_-_431249 | 1.69 |
ENSDART00000083827
|
hs3st3l
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3-like |
| chr10_-_29101715 | 1.68 |
ENSDART00000149674
ENSDART00000171194 ENSDART00000192019 |
si:ch211-103f14.3
|
si:ch211-103f14.3 |
| chr10_+_158590 | 1.68 |
ENSDART00000081982
|
KCNJ15
|
potassium voltage-gated channel subfamily J member 15 |
| chr8_-_33154677 | 1.64 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr11_+_6159595 | 1.64 |
ENSDART00000178367
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr9_-_7421135 | 1.61 |
ENSDART00000144600
|
bin1a
|
bridging integrator 1a |
| chr2_+_37110504 | 1.60 |
ENSDART00000132794
ENSDART00000042974 |
slc1a8b
|
solute carrier family 1 (glutamate transporter), member 8b |
| chr3_+_3810919 | 1.59 |
ENSDART00000056035
|
FQ311927.1
|
|
| chr14_+_25817628 | 1.58 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr12_-_31461219 | 1.54 |
ENSDART00000148954
|
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr12_+_6195191 | 1.43 |
ENSDART00000043236
ENSDART00000186420 |
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr23_+_36601984 | 1.42 |
ENSDART00000128598
|
igfbp6b
|
insulin-like growth factor binding protein 6b |
| chr6_+_55277419 | 1.40 |
ENSDART00000083670
|
CABZ01041604.1
|
|
| chr2_+_38608290 | 1.40 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr23_+_26946744 | 1.37 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr14_-_2196267 | 1.34 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr15_-_24869826 | 1.33 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr16_-_47483142 | 1.32 |
ENSDART00000147072
|
cthrc1b
|
collagen triple helix repeat containing 1b |
| chr20_-_47732703 | 1.31 |
ENSDART00000193975
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr24_+_32472155 | 1.31 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr18_-_21177674 | 1.27 |
ENSDART00000060175
|
si:dkey-12e7.4
|
si:dkey-12e7.4 |
| chr15_-_1941042 | 1.26 |
ENSDART00000112946
ENSDART00000155504 |
dock10
|
dedicator of cytokinesis 10 |
| chr17_+_28533102 | 1.25 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr12_-_7807298 | 1.23 |
ENSDART00000191355
|
ank3b
|
ankyrin 3b |
| chr24_+_7800486 | 1.22 |
ENSDART00000145504
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr4_+_16323970 | 1.22 |
ENSDART00000190651
|
BX322608.1
|
|
| chr5_-_5831037 | 1.21 |
ENSDART00000112856
|
zmp:0000000846
|
zmp:0000000846 |
| chr8_-_27515540 | 1.18 |
ENSDART00000146132
|
si:ch211-254n4.3
|
si:ch211-254n4.3 |
| chr18_+_45504362 | 1.13 |
ENSDART00000140089
|
cngb1a
|
cyclic nucleotide gated channel beta 1a |
| chr8_-_3716734 | 1.12 |
ENSDART00000172966
|
bicdl1
|
BICD family like cargo adaptor 1 |
| chr9_-_22831836 | 1.09 |
ENSDART00000142585
|
neb
|
nebulin |
| chr25_-_31739309 | 1.08 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr15_+_16521785 | 1.08 |
ENSDART00000062191
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr15_+_37559570 | 1.08 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr3_-_47038481 | 1.07 |
ENSDART00000193260
|
CR392045.1
|
|
| chr6_-_56111829 | 1.03 |
ENSDART00000154397
|
gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr17_+_37645633 | 1.03 |
ENSDART00000085500
|
tmem229b
|
transmembrane protein 229B |
| chr3_+_16722014 | 1.02 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr20_+_27020201 | 1.02 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr7_+_66822229 | 1.01 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr1_-_40994259 | 1.01 |
ENSDART00000101562
|
adra2c
|
adrenoceptor alpha 2C |
| chr7_-_56793739 | 1.00 |
ENSDART00000082842
|
si:ch211-146m13.3
|
si:ch211-146m13.3 |
| chr7_-_41693004 | 1.00 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr20_-_25669813 | 0.98 |
ENSDART00000153118
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr9_-_21912227 | 0.98 |
ENSDART00000145576
|
lmo7a
|
LIM domain 7a |
| chr14_-_27289042 | 0.97 |
ENSDART00000159727
|
pcdh11
|
protocadherin 11 |
| chr25_+_4541211 | 0.97 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr12_+_45677293 | 0.95 |
ENSDART00000152850
ENSDART00000153047 |
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr23_-_35064785 | 0.93 |
ENSDART00000172240
|
BX294434.1
|
|
| chr12_+_48744016 | 0.93 |
ENSDART00000175518
|
LO017648.1
|
|
| chr1_-_44701313 | 0.93 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr1_+_41690402 | 0.91 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr15_-_23908779 | 0.87 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr16_-_13109749 | 0.85 |
ENSDART00000142610
|
prkcg
|
protein kinase C, gamma |
| chr15_+_35718354 | 0.83 |
ENSDART00000110003
|
nyap2a
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2a |
| chr14_-_858985 | 0.82 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr1_-_17675037 | 0.81 |
ENSDART00000142689
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr9_+_41612642 | 0.80 |
ENSDART00000138473
|
spegb
|
SPEG complex locus b |
| chr2_-_30987538 | 0.80 |
ENSDART00000148157
|
emilin2a
|
elastin microfibril interfacer 2a |
| chr25_+_35212919 | 0.80 |
ENSDART00000180127
|
ano5a
|
anoctamin 5a |
| chr4_+_20263097 | 0.80 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr1_+_19708508 | 0.79 |
ENSDART00000054581
ENSDART00000131206 |
march1
|
membrane-associated ring finger (C3HC4) 1 |
| chr19_+_56351 | 0.79 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr23_+_26946429 | 0.79 |
ENSDART00000185564
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr20_-_6936488 | 0.78 |
ENSDART00000168651
|
adcy1a
|
adenylate cyclase 1a |
| chr25_-_1235457 | 0.77 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr15_-_47956388 | 0.77 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr25_+_29688671 | 0.77 |
ENSDART00000073478
|
brd1b
|
bromodomain containing 1b |
| chr20_-_53321499 | 0.76 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr24_-_31843173 | 0.75 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr24_+_26402110 | 0.73 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr9_-_39547907 | 0.72 |
ENSDART00000163635
|
erbb4b
|
erb-b2 receptor tyrosine kinase 4b |
| chr9_+_56443416 | 0.72 |
ENSDART00000167947
|
CABZ01079480.1
|
|
| chr22_-_27136451 | 0.70 |
ENSDART00000153589
|
si:dkey-16m19.1
|
si:dkey-16m19.1 |
| chr8_+_8532407 | 0.68 |
ENSDART00000169276
ENSDART00000138993 |
grm6a
|
glutamate receptor, metabotropic 6a |
| chr18_+_27515640 | 0.67 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr3_-_3496738 | 0.66 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr23_+_14269311 | 0.66 |
ENSDART00000170414
|
CR354537.1
|
|
| chr25_+_35706493 | 0.65 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr10_-_28483672 | 0.64 |
ENSDART00000139994
|
bbx
|
bobby sox homolog (Drosophila) |
| chr5_-_38063585 | 0.63 |
ENSDART00000164947
|
si:dkey-111e8.5
|
si:dkey-111e8.5 |
| chr4_+_5193198 | 0.63 |
ENSDART00000067388
|
fgf23
|
fibroblast growth factor 23 |
| chr19_-_35596207 | 0.63 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr19_+_58954 | 0.62 |
ENSDART00000162379
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr6_+_45692026 | 0.60 |
ENSDART00000164759
|
cntn4
|
contactin 4 |
| chr13_-_47800342 | 0.59 |
ENSDART00000121817
|
fbln7
|
fibulin 7 |
| chr12_-_46176115 | 0.59 |
ENSDART00000152848
|
si:ch211-226h7.8
|
si:ch211-226h7.8 |
| chr20_+_18209895 | 0.57 |
ENSDART00000111063
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr22_-_32360464 | 0.57 |
ENSDART00000148886
|
pcbp4
|
poly(rC) binding protein 4 |
| chr1_-_191971 | 0.55 |
ENSDART00000164091
ENSDART00000159237 |
grtp1a
|
growth hormone regulated TBC protein 1a |
| chr6_+_58406014 | 0.55 |
ENSDART00000044241
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr14_-_6727717 | 0.54 |
ENSDART00000166979
|
si:dkeyp-44a8.4
|
si:dkeyp-44a8.4 |
| chr18_+_34225520 | 0.53 |
ENSDART00000126115
|
v2rl1
|
vomeronasal 2 receptor, l1 |
| chr13_-_13751017 | 0.53 |
ENSDART00000180182
|
ky
|
kyphoscoliosis peptidase |
| chr15_-_5157572 | 0.53 |
ENSDART00000174192
|
or128-10
|
odorant receptor, family E, subfamily 128, member 10 |
| chr7_-_24005268 | 0.53 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr14_-_41535822 | 0.52 |
ENSDART00000149407
|
itga6l
|
integrin, alpha 6, like |
| chr15_-_34214440 | 0.52 |
ENSDART00000167052
|
etv1
|
ets variant 1 |
| chr13_-_4018888 | 0.52 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr4_+_4272622 | 0.52 |
ENSDART00000164810
|
ano2
|
anoctamin 2 |
| chr9_-_27442339 | 0.52 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr7_+_11459235 | 0.50 |
ENSDART00000159611
|
il16
|
interleukin 16 |
| chr21_-_45186431 | 0.48 |
ENSDART00000193449
|
si:ch73-269m14.2
|
si:ch73-269m14.2 |
| chr12_+_31460877 | 0.48 |
ENSDART00000031154
|
gucy2g
|
guanylate cyclase 2g |
| chr17_+_45454943 | 0.47 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr24_+_1294176 | 0.47 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr2_-_52020049 | 0.46 |
ENSDART00000128198
ENSDART00000135331 |
scn12aa
|
sodium channel, voltage gated, type XII, alpha a |
| chr1_-_46989650 | 0.46 |
ENSDART00000129072
ENSDART00000150507 |
si:dkey-22n8.3
|
si:dkey-22n8.3 |
| chr11_+_7214353 | 0.46 |
ENSDART00000156764
|
nwd1
|
NACHT and WD repeat domain containing 1 |
| chr23_-_36418059 | 0.44 |
ENSDART00000135232
|
znf740b
|
zinc finger protein 740b |
| chr5_+_24287927 | 0.43 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr15_+_28268135 | 0.43 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr14_+_41887070 | 0.43 |
ENSDART00000173264
|
slc7a11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr25_-_24074500 | 0.43 |
ENSDART00000040410
|
th
|
tyrosine hydroxylase |
| chr22_+_28446365 | 0.43 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr20_+_38201644 | 0.41 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr13_+_12045758 | 0.41 |
ENSDART00000079398
ENSDART00000165467 ENSDART00000165880 |
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr2_-_37478418 | 0.40 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr21_-_14775724 | 0.39 |
ENSDART00000043239
|
glulc
|
glutamate-ammonia ligase (glutamine synthase) c |
| chr12_-_33659328 | 0.39 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr7_+_73807504 | 0.38 |
ENSDART00000166244
|
si:ch73-252p3.1
|
si:ch73-252p3.1 |
| chr1_-_58880613 | 0.36 |
ENSDART00000188136
|
CABZ01084501.2
|
|
| chr3_+_26081343 | 0.36 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr12_-_31484677 | 0.35 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr5_+_37343643 | 0.34 |
ENSDART00000163993
ENSDART00000164459 ENSDART00000167006 ENSDART00000128122 ENSDART00000183720 |
klhl13
|
kelch-like family member 13 |
| chr3_-_11203689 | 0.33 |
ENSDART00000166328
|
BX323882.1
|
|
| chr18_-_210478 | 0.32 |
ENSDART00000136693
|
tm2d3
|
TM2 domain containing 3 |
| chr7_-_29223614 | 0.32 |
ENSDART00000173598
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr9_-_45149695 | 0.29 |
ENSDART00000162033
|
CU467837.1
|
|
| chr3_+_46315016 | 0.27 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr25_+_2415131 | 0.27 |
ENSDART00000162210
|
zmp:0000000932
|
zmp:0000000932 |
| chr21_-_11855828 | 0.26 |
ENSDART00000081666
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr4_-_20115570 | 0.26 |
ENSDART00000134528
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
| chr5_-_29112956 | 0.26 |
ENSDART00000132726
|
whrnb
|
whirlin b |
| chr17_-_15528597 | 0.24 |
ENSDART00000150232
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr9_+_44430974 | 0.24 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr3_-_54846444 | 0.23 |
ENSDART00000074010
|
ubald1b
|
UBA-like domain containing 1b |
| chr19_+_5651325 | 0.23 |
ENSDART00000082067
|
ft1
|
alpha(1,3)fucosyltransferase gene 1 |
| chr15_-_5245726 | 0.23 |
ENSDART00000174079
ENSDART00000174041 |
or128-1
|
odorant receptor, family E, subfamily 128, member 1 |
| chr7_-_44604821 | 0.23 |
ENSDART00000148967
|
tk2
|
thymidine kinase 2, mitochondrial |
| chr21_-_11820379 | 0.21 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr12_-_43693192 | 0.21 |
ENSDART00000172284
|
CU914622.1
|
|
| chr18_-_7112882 | 0.21 |
ENSDART00000176292
|
si:dkey-88e18.8
|
si:dkey-88e18.8 |
| chr11_+_6739433 | 0.20 |
ENSDART00000163397
|
pde4cb
|
phosphodiesterase 4C, cAMP-specific b |
| chr21_-_14773692 | 0.20 |
ENSDART00000142145
|
glulc
|
glutamate-ammonia ligase (glutamine synthase) c |
| chr21_-_11791909 | 0.20 |
ENSDART00000180893
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr22_+_20710434 | 0.19 |
ENSDART00000135521
|
peak3
|
PEAK family member 3 |
| chr8_-_19051906 | 0.16 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr6_+_42475730 | 0.16 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr20_-_49657134 | 0.15 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr17_+_30591287 | 0.14 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr3_+_32416948 | 0.14 |
ENSDART00000157324
ENSDART00000154267 ENSDART00000186094 ENSDART00000155860 ENSDART00000156986 |
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr22_+_36914636 | 0.14 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr10_+_43039947 | 0.14 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr10_-_32639939 | 0.13 |
ENSDART00000126952
ENSDART00000099853 |
umodl1
|
uromodulin-like 1 |
| chr6_+_11850359 | 0.12 |
ENSDART00000109552
ENSDART00000188139 ENSDART00000181499 ENSDART00000178269 |
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr22_-_30881738 | 0.12 |
ENSDART00000059965
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr1_-_54191626 | 0.11 |
ENSDART00000062941
|
nthl1
|
nth-like DNA glycosylase 1 |
| chr16_-_26125304 | 0.11 |
ENSDART00000176517
|
lipeb
|
lipase, hormone-sensitive b |
| chr21_-_15929041 | 0.10 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr6_+_40794015 | 0.10 |
ENSDART00000144479
|
gata2b
|
GATA binding protein 2b |
| chr21_-_26558268 | 0.10 |
ENSDART00000065390
|
CR631122.1
|
|
| chr4_-_9579299 | 0.09 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr7_-_52334840 | 0.09 |
ENSDART00000174173
|
CR938716.1
|
|
| chr11_+_19060079 | 0.08 |
ENSDART00000158123
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr23_-_1144425 | 0.08 |
ENSDART00000098271
|
psmb11b
|
proteasome subunit beta11b |
| chr8_-_34762163 | 0.08 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr15_-_5207805 | 0.08 |
ENSDART00000174046
|
or128-7
|
odorant receptor, family E, subfamily 128, member 7 |
| chr15_+_31515976 | 0.08 |
ENSDART00000156471
|
tex26
|
testis expressed 26 |
| chr1_-_8963484 | 0.08 |
ENSDART00000188035
|
socs1b
|
suppressor of cytokine signaling 1b |
| chr22_+_2844865 | 0.07 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr21_+_21685228 | 0.06 |
ENSDART00000142844
|
or125-4
|
odorant receptor, family E, subfamily 125, member 4 |
| chr21_+_26389391 | 0.06 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr18_+_20031046 | 0.06 |
ENSDART00000132656
|
rxfp3.3a3
|
relaxin/insulin-like family peptide receptor 3.3a3 |
| chr10_-_27199135 | 0.06 |
ENSDART00000189511
ENSDART00000180314 |
auts2a
|
autism susceptibility candidate 2a |
| chr7_+_34487833 | 0.05 |
ENSDART00000173854
|
cln6a
|
CLN6, transmembrane ER protein a |
| chr19_-_2421793 | 0.05 |
ENSDART00000180238
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr25_+_9003230 | 0.05 |
ENSDART00000180330
ENSDART00000142917 |
rag1
|
recombination activating gene 1 |
| chr6_+_58508420 | 0.05 |
ENSDART00000153740
|
chrna4a
|
cholinergic receptor, nicotinic, alpha 4a |
| chr24_+_17334682 | 0.04 |
ENSDART00000018868
|
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr8_+_48858132 | 0.03 |
ENSDART00000124737
ENSDART00000079644 |
tp73
|
tumor protein p73 |
| chr2_-_23603061 | 0.03 |
ENSDART00000132072
|
si:dkey-58b18.9
|
si:dkey-58b18.9 |
| chr23_+_19814371 | 0.03 |
ENSDART00000182897
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr4_-_58964138 | 0.03 |
ENSDART00000150259
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr3_+_13559199 | 0.02 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ebf3a-1+si:ch211-51e8.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.5 | 1.5 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.4 | 1.2 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.4 | 2.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.3 | 1.0 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.3 | 1.7 | GO:0051701 | interaction with host(GO:0051701) |
| 0.3 | 1.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.2 | 1.0 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 0.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 1.0 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.2 | 0.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 0.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 1.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 1.4 | GO:0072502 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 1.7 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.4 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.1 | 1.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 3.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.4 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 1.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.5 | GO:0050930 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 1.0 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 2.1 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.6 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.0 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 3.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.8 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.5 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) cAMP metabolic process(GO:0046058) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 1.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 4.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 2.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.9 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.3 | 2.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 1.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 1.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 1.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 2.2 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.2 | 1.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.7 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 3.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 1.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 0.5 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 1.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.4 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 1.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 1.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.6 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 1.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 1.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 3.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 3.1 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 3.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.4 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 1.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 3.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.6 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 2.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |