Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for e2f2_e2f5
Z-value: 4.47
Transcription factors associated with e2f2_e2f5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
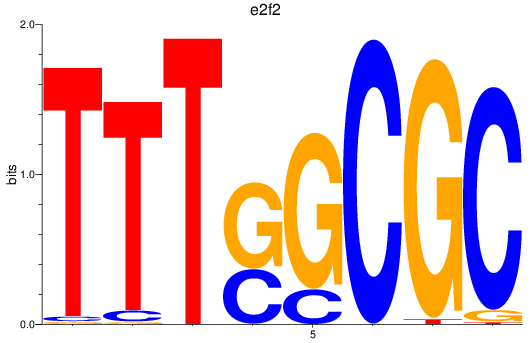
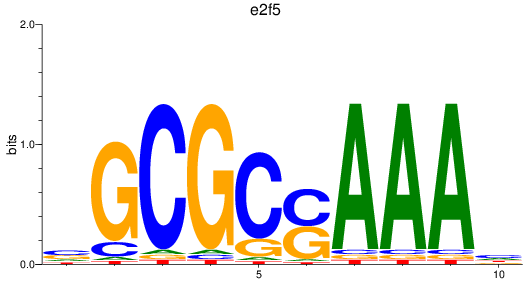
e2f2
|
ENSDARG00000079233 | si_ch211-160f23.5 |
|
e2f5
|
ENSDARG00000038812 | E2F transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F2 | dr11_v1_chr17_-_27266053_27266053 | 0.93 | 9.7e-09 | Click! |
| e2f5 | dr11_v1_chr2_-_31686353_31686403 | 0.14 | 5.7e-01 | Click! |
Activity profile of e2f2_e2f5 motif
Sorted Z-values of e2f2_e2f5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_39344889 | 20.76 |
ENSDART00000009164
|
esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr20_+_34770197 | 20.18 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr19_-_47571797 | 19.18 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr13_+_8255106 | 16.79 |
ENSDART00000080465
|
hells
|
helicase, lymphoid specific |
| chr9_-_2892250 | 15.73 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr3_-_54607166 | 15.62 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr22_-_4439311 | 12.94 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr11_-_11792766 | 12.83 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr24_-_35561672 | 11.99 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr4_-_2545310 | 11.87 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr2_-_29923403 | 11.77 |
ENSDART00000144672
|
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr3_+_25999477 | 11.57 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr12_+_9499993 | 11.39 |
ENSDART00000135871
|
dnajc9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr2_-_37837056 | 11.27 |
ENSDART00000158179
ENSDART00000160317 ENSDART00000171409 |
mettl17
|
methyltransferase like 17 |
| chr7_+_55518519 | 10.85 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr24_+_11908480 | 10.80 |
ENSDART00000024224
|
fen1
|
flap structure-specific endonuclease 1 |
| chr13_-_13030851 | 10.79 |
ENSDART00000009499
|
nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr25_-_35095129 | 10.76 |
ENSDART00000099866
ENSDART00000099868 |
kif15
|
kinesin family member 15 |
| chr23_+_31815423 | 10.43 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr2_-_29923630 | 10.43 |
ENSDART00000158844
ENSDART00000031130 |
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr19_-_29294457 | 10.39 |
ENSDART00000130815
ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr3_+_26244353 | 10.17 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr24_+_11908833 | 10.02 |
ENSDART00000178622
|
fen1
|
flap structure-specific endonuclease 1 |
| chr20_-_29499363 | 9.87 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr24_+_23840821 | 9.49 |
ENSDART00000128595
ENSDART00000127188 |
pola1
|
polymerase (DNA directed), alpha 1 |
| chr8_-_4760723 | 9.48 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr2_+_37836821 | 9.43 |
ENSDART00000143203
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr1_+_19538299 | 9.37 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr12_+_9499742 | 9.27 |
ENSDART00000044150
ENSDART00000136354 |
dnajc9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr7_+_34236238 | 9.12 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr21_+_22558187 | 9.09 |
ENSDART00000167599
|
chek1
|
checkpoint kinase 1 |
| chr7_-_6464225 | 9.04 |
ENSDART00000130760
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr22_-_3152357 | 9.02 |
ENSDART00000170983
|
lmnb2
|
lamin B2 |
| chr17_+_8799451 | 8.86 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr18_-_16924221 | 8.71 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr6_+_33931740 | 8.61 |
ENSDART00000130492
ENSDART00000151213 |
orc1
|
origin recognition complex, subunit 1 |
| chr9_+_22780901 | 8.60 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
| chr2_+_38055529 | 8.50 |
ENSDART00000145642
|
si:rp71-1g18.1
|
si:rp71-1g18.1 |
| chr6_+_12968101 | 8.46 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr19_-_10214264 | 8.39 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr6_+_7533601 | 8.34 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr3_-_50046004 | 8.33 |
ENSDART00000109544
|
si:ch1073-100f3.2
|
si:ch1073-100f3.2 |
| chr19_-_47571456 | 8.20 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr19_+_48060464 | 8.06 |
ENSDART00000123163
|
zgc:85936
|
zgc:85936 |
| chr17_+_8799661 | 8.06 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr5_+_12743640 | 7.99 |
ENSDART00000081411
|
pole
|
polymerase (DNA directed), epsilon |
| chr9_-_2892045 | 7.93 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr7_+_24023653 | 7.78 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr7_+_41812636 | 7.71 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr6_-_34838397 | 7.60 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr21_-_26490186 | 7.48 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr1_+_29858032 | 7.35 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr2_-_38363017 | 7.28 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr1_-_8553165 | 7.11 |
ENSDART00000135197
ENSDART00000054981 |
zgc:112980
|
zgc:112980 |
| chr3_+_22335030 | 7.07 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr7_+_41812817 | 7.05 |
ENSDART00000174165
|
orc6
|
origin recognition complex, subunit 6 |
| chr3_-_26244256 | 7.01 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr12_-_33817114 | 7.01 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr11_-_309420 | 6.95 |
ENSDART00000173185
|
poc1a
|
POC1 centriolar protein A |
| chr4_+_18441988 | 6.92 |
ENSDART00000040827
|
ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr5_+_34549365 | 6.88 |
ENSDART00000009500
|
aif1l
|
allograft inflammatory factor 1-like |
| chr6_-_51573975 | 6.77 |
ENSDART00000073865
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr20_-_29498178 | 6.70 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr8_-_40327397 | 6.70 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr20_-_38827623 | 6.68 |
ENSDART00000153310
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr25_+_36327034 | 6.64 |
ENSDART00000073452
|
zgc:110216
|
zgc:110216 |
| chr16_+_41015163 | 6.61 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr8_+_52415603 | 6.56 |
ENSDART00000021604
ENSDART00000191424 |
gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr1_-_59313465 | 6.52 |
ENSDART00000158067
ENSDART00000159419 |
txndc11
|
thioredoxin domain containing 11 |
| chr19_+_9455218 | 6.30 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr15_-_26538989 | 6.28 |
ENSDART00000032880
|
rpa1
|
replication protein A1 |
| chr17_+_4325693 | 6.22 |
ENSDART00000154264
|
mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr2_+_21000334 | 6.19 |
ENSDART00000062563
ENSDART00000147809 |
rreb1b
|
ras responsive element binding protein 1b |
| chr3_-_35865040 | 6.16 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr19_-_2822372 | 6.16 |
ENSDART00000109130
ENSDART00000122385 |
recql4
|
RecQ helicase-like 4 |
| chr22_+_1930589 | 5.97 |
ENSDART00000159807
|
znf1153
|
zinc finger protein 1153 |
| chr4_+_69888328 | 5.97 |
ENSDART00000170163
|
si:ch211-145h19.5
|
si:ch211-145h19.5 |
| chr2_+_44512324 | 5.92 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr5_+_34549845 | 5.92 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr6_-_33931696 | 5.84 |
ENSDART00000057290
|
prpf38a
|
pre-mRNA processing factor 38A |
| chr23_-_18913032 | 5.73 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr7_+_41812190 | 5.68 |
ENSDART00000113732
ENSDART00000174137 |
orc6
|
origin recognition complex, subunit 6 |
| chr20_-_23440955 | 5.60 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr25_-_13403726 | 5.59 |
ENSDART00000056723
|
gins3
|
GINS complex subunit 3 |
| chr16_-_43335914 | 5.57 |
ENSDART00000111963
|
atad2
|
ATPase family, AAA domain containing 2 |
| chr17_-_27266053 | 5.55 |
ENSDART00000110903
|
E2F2
|
si:ch211-160f23.5 |
| chr7_+_71586485 | 5.54 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr6_+_32046202 | 5.54 |
ENSDART00000156552
|
si:dkey-148h10.5
|
si:dkey-148h10.5 |
| chr20_-_9199721 | 5.52 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr14_-_15956657 | 5.45 |
ENSDART00000169197
|
flt4
|
fms-related tyrosine kinase 4 |
| chr16_+_40575742 | 5.42 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr5_+_23136544 | 5.39 |
ENSDART00000003428
ENSDART00000109340 ENSDART00000171039 ENSDART00000178821 |
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr20_+_43648369 | 5.31 |
ENSDART00000187930
ENSDART00000017269 |
parp1
|
poly (ADP-ribose) polymerase 1 |
| chr24_-_21258945 | 5.19 |
ENSDART00000111025
|
boc
|
BOC cell adhesion associated, oncogene regulated |
| chr4_+_70556298 | 5.13 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr21_-_27413294 | 5.12 |
ENSDART00000131646
|
slc29a2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr2_+_37837249 | 5.10 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr12_+_9183626 | 5.04 |
ENSDART00000020192
|
tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr5_-_72324371 | 4.99 |
ENSDART00000006380
|
tbx3a
|
T-box 3a |
| chr23_+_43950674 | 4.97 |
ENSDART00000167813
|
corin
|
corin, serine peptidase |
| chr16_-_21140097 | 4.96 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr19_-_20199167 | 4.88 |
ENSDART00000155527
|
si:ch211-155k24.9
|
si:ch211-155k24.9 |
| chr19_-_14191592 | 4.86 |
ENSDART00000164594
|
tbxta
|
T-box transcription factor Ta |
| chr12_+_35650321 | 4.86 |
ENSDART00000190446
|
BX255898.1
|
|
| chr10_+_21434649 | 4.86 |
ENSDART00000193938
ENSDART00000064558 |
etf1b
|
eukaryotic translation termination factor 1b |
| chr3_+_37112693 | 4.84 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr7_-_40657831 | 4.78 |
ENSDART00000084153
|
nom1
|
nucleolar protein with MIF4G domain 1 |
| chr8_+_10304981 | 4.77 |
ENSDART00000160766
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr2_-_44344321 | 4.68 |
ENSDART00000084174
|
lig1
|
ligase I, DNA, ATP-dependent |
| chr7_+_1550966 | 4.65 |
ENSDART00000177863
ENSDART00000126840 |
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr17_-_22573311 | 4.62 |
ENSDART00000141523
ENSDART00000140022 ENSDART00000079390 ENSDART00000188644 |
exo1
|
exonuclease 1 |
| chr20_+_41801913 | 4.61 |
ENSDART00000139805
|
mcm9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chr19_-_20777351 | 4.60 |
ENSDART00000019206
|
ngly1
|
N-glycanase 1 |
| chr23_-_17003533 | 4.58 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr19_-_31007417 | 4.49 |
ENSDART00000048144
|
rbbp4
|
retinoblastoma binding protein 4 |
| chr23_-_45339439 | 4.47 |
ENSDART00000148726
|
ccdc171
|
coiled-coil domain containing 171 |
| chr24_-_37338739 | 4.45 |
ENSDART00000146844
|
tsr3
|
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr2_-_44777592 | 4.45 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr4_-_9909371 | 4.43 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr5_-_19014589 | 4.36 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr20_-_29633507 | 4.35 |
ENSDART00000040292
|
cpsf3
|
cleavage and polyadenylation specific factor 3 |
| chr22_-_7006974 | 4.29 |
ENSDART00000133143
|
gpd1b
|
glycerol-3-phosphate dehydrogenase 1b |
| chr15_-_14884332 | 4.28 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr10_+_33588715 | 4.16 |
ENSDART00000051198
|
mis18a
|
MIS18 kinetochore protein A |
| chr1_-_42289704 | 4.13 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr8_-_25761544 | 4.05 |
ENSDART00000078152
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr13_+_9612395 | 4.04 |
ENSDART00000136689
ENSDART00000138362 |
slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr13_-_35908275 | 4.04 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr25_-_32363341 | 4.03 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr9_+_29671553 | 3.99 |
ENSDART00000015377
|
rnaseh2b
|
ribonuclease H2, subunit B |
| chr4_-_5831522 | 3.88 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr5_+_29159777 | 3.88 |
ENSDART00000174702
ENSDART00000037891 |
dpp7
|
dipeptidyl-peptidase 7 |
| chr7_+_26534131 | 3.88 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr5_-_54395488 | 3.83 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr6_-_1762191 | 3.82 |
ENSDART00000167928
|
orc4
|
origin recognition complex, subunit 4 |
| chr5_+_22969651 | 3.75 |
ENSDART00000089992
ENSDART00000145477 |
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr22_-_5822147 | 3.72 |
ENSDART00000011076
|
cers5
|
ceramide synthase 5 |
| chr19_-_2582858 | 3.68 |
ENSDART00000113829
|
cdca7b
|
cell division cycle associated 7b |
| chr2_-_37837472 | 3.67 |
ENSDART00000165347
|
mettl17
|
methyltransferase like 17 |
| chr18_-_20532339 | 3.66 |
ENSDART00000060291
|
ighmbp2
|
immunoglobulin mu binding protein 2 |
| chr7_+_40884012 | 3.58 |
ENSDART00000149395
|
shha
|
sonic hedgehog a |
| chr11_+_18216404 | 3.58 |
ENSDART00000086437
|
tmcc1b
|
transmembrane and coiled-coil domain family 1b |
| chr14_-_30918662 | 3.58 |
ENSDART00000176631
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr12_-_3962372 | 3.53 |
ENSDART00000016791
|
eif3c
|
eukaryotic translation initiation factor 3, subunit C |
| chr15_-_23784600 | 3.51 |
ENSDART00000059354
|
rad1
|
RAD1 homolog (S. pombe) |
| chr10_-_21362071 | 3.51 |
ENSDART00000125167
|
avd
|
avidin |
| chr7_+_24814866 | 3.47 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr24_+_17345521 | 3.41 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr1_-_20068155 | 3.38 |
ENSDART00000102993
|
mettl14
|
methyltransferase like 14 |
| chr21_+_30721733 | 3.35 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr9_+_28232522 | 3.34 |
ENSDART00000031761
|
fzd5
|
frizzled class receptor 5 |
| chr12_+_17927871 | 3.32 |
ENSDART00000172062
|
trrap
|
transformation/transcription domain-associated protein |
| chr7_+_71535045 | 3.30 |
ENSDART00000047069
|
tyms
|
thymidylate synthetase |
| chr1_+_26677406 | 3.30 |
ENSDART00000183427
ENSDART00000180366 ENSDART00000181997 |
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr2_-_47957673 | 3.28 |
ENSDART00000056305
|
fzd8b
|
frizzled class receptor 8b |
| chr13_+_48358467 | 3.27 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr5_+_61475451 | 3.18 |
ENSDART00000163444
|
lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr25_-_12824656 | 3.17 |
ENSDART00000171801
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr19_+_25465025 | 3.17 |
ENSDART00000018553
|
rpa3
|
replication protein A3 |
| chr1_-_54622227 | 3.10 |
ENSDART00000049010
|
tekt4
|
tektin 4 |
| chr7_-_7797654 | 3.07 |
ENSDART00000084503
ENSDART00000192779 ENSDART00000173079 |
trmt10b
|
tRNA methyltransferase 10B |
| chr1_+_9199031 | 3.05 |
ENSDART00000092058
ENSDART00000182771 |
chtf18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
| chr18_+_41561285 | 3.04 |
ENSDART00000169621
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr13_+_12606821 | 2.99 |
ENSDART00000140096
ENSDART00000145136 |
metap1
|
methionyl aminopeptidase 1 |
| chr14_+_22129096 | 2.97 |
ENSDART00000132514
|
ccng1
|
cyclin G1 |
| chr25_-_35140746 | 2.94 |
ENSDART00000129969
|
si:ch211-113a14.19
|
si:ch211-113a14.19 |
| chr22_+_2819613 | 2.93 |
ENSDART00000131234
|
si:dkey-20i20.3
|
si:dkey-20i20.3 |
| chr17_-_29224908 | 2.92 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr25_-_34740627 | 2.91 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr16_+_28728347 | 2.91 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr3_+_53116172 | 2.89 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr18_-_30499489 | 2.85 |
ENSDART00000033746
|
gins2
|
GINS complex subunit 2 |
| chr13_-_36844945 | 2.80 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr24_-_38657683 | 2.80 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr22_-_20419660 | 2.79 |
ENSDART00000105520
|
pias4a
|
protein inhibitor of activated STAT, 4a |
| chr1_+_26676758 | 2.79 |
ENSDART00000152299
|
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr21_-_23017478 | 2.79 |
ENSDART00000024309
|
rb1
|
retinoblastoma 1 |
| chr1_-_59216197 | 2.78 |
ENSDART00000062426
|
lpar2b
|
lysophosphatidic acid receptor 2b |
| chr15_+_23784842 | 2.76 |
ENSDART00000192889
ENSDART00000138375 |
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr9_-_30576522 | 2.76 |
ENSDART00000101085
|
morc3a
|
MORC family CW-type zinc finger 3a |
| chr24_-_41478917 | 2.76 |
ENSDART00000192192
|
CABZ01084131.1
|
|
| chr17_-_49407091 | 2.76 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr16_+_7154266 | 2.73 |
ENSDART00000168830
|
bmper
|
BMP binding endothelial regulator |
| chr8_+_30112655 | 2.72 |
ENSDART00000099027
|
fancc
|
Fanconi anemia, complementation group C |
| chr4_+_77933084 | 2.70 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr22_+_1940595 | 2.69 |
ENSDART00000163506
|
znf1167
|
zinc finger protein 1167 |
| chr6_+_52242139 | 2.69 |
ENSDART00000041549
|
zc3h10
|
zinc finger CCCH-type containing 10 |
| chr13_-_35907768 | 2.68 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr5_-_58832332 | 2.65 |
ENSDART00000161230
|
arhgef12b
|
Rho guanine nucleotide exchange factor (GEF) 12b |
| chr15_-_1584137 | 2.64 |
ENSDART00000056772
|
trim59
|
tripartite motif containing 59 |
| chr21_-_19915757 | 2.62 |
ENSDART00000164317
|
eri1
|
exoribonuclease 1 |
| chr13_+_5013572 | 2.59 |
ENSDART00000162425
|
psap
|
prosaposin |
| chr14_+_24241241 | 2.57 |
ENSDART00000022377
|
nkx2.5
|
NK2 homeobox 5 |
| chr9_+_35014728 | 2.56 |
ENSDART00000100700
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr20_-_54245256 | 2.55 |
ENSDART00000170482
|
prpf39
|
PRP39 pre-mRNA processing factor 39 homolog (yeast) |
| chr25_+_36339867 | 2.54 |
ENSDART00000152195
|
si:ch211-113a14.18
|
si:ch211-113a14.18 |
| chr25_+_35062353 | 2.53 |
ENSDART00000089844
|
zgc:113983
|
zgc:113983 |
| chr16_+_41015781 | 2.51 |
ENSDART00000124543
|
dek
|
DEK proto-oncogene |
| chr5_+_24179307 | 2.51 |
ENSDART00000051552
|
mpdu1a
|
mannose-P-dolichol utilization defect 1a |
| chr22_+_1947494 | 2.49 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f2_e2f5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 20.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 6.8 | 47.3 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 4.5 | 45.3 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 4.3 | 47.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 3.6 | 25.5 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 3.6 | 10.8 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 3.4 | 43.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 3.0 | 41.4 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 2.8 | 11.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 2.7 | 8.0 | GO:0006272 | leading strand elongation(GO:0006272) |
| 2.4 | 7.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 2.4 | 11.9 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 2.2 | 6.7 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 1.9 | 24.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 1.7 | 10.4 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 1.7 | 6.7 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 1.7 | 8.3 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 1.7 | 19.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 1.6 | 9.7 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.5 | 1.5 | GO:0043111 | replication fork arrest(GO:0043111) |
| 1.4 | 12.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.3 | 5.4 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.3 | 10.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 1.2 | 6.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.2 | 4.9 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 1.1 | 4.6 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 1.1 | 3.4 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 1.1 | 3.3 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 1.1 | 5.5 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 1.1 | 4.3 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 1.1 | 7.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 1.0 | 20.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 1.0 | 4.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.0 | 7.0 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 1.0 | 5.0 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.0 | 4.9 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 1.0 | 2.9 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.9 | 6.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.9 | 3.6 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.9 | 4.4 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.9 | 2.6 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.9 | 5.1 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.8 | 2.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.8 | 3.2 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.8 | 13.3 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.7 | 3.0 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.7 | 4.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.7 | 5.9 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.7 | 2.8 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.7 | 2.8 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.7 | 3.4 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.7 | 3.4 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.7 | 25.4 | GO:0048538 | thymus development(GO:0048538) |
| 0.6 | 12.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.6 | 10.8 | GO:0032508 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.6 | 1.9 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.6 | 2.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.6 | 3.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.6 | 2.3 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.6 | 2.8 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.5 | 7.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 2.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.5 | 2.1 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.5 | 4.7 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.5 | 10.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.5 | 3.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.5 | 4.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.5 | 9.6 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.4 | 9.4 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.4 | 2.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 7.3 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.4 | 1.9 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.4 | 2.9 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.4 | 1.4 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.4 | 1.4 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.4 | 3.9 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.3 | 0.7 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.3 | 1.6 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.3 | 1.9 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 1.8 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.3 | 1.2 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) immunoglobulin V(D)J recombination(GO:0033152) T cell receptor V(D)J recombination(GO:0033153) |
| 0.3 | 5.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.3 | 2.7 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.3 | 7.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.3 | 9.1 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.3 | 8.6 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.3 | 2.3 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.3 | 1.5 | GO:0060969 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) negative regulation of gene silencing(GO:0060969) 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 5.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.2 | 1.4 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.2 | 3.1 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.2 | 0.2 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 5.0 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.2 | 7.9 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.2 | 1.5 | GO:0014004 | microglia differentiation(GO:0014004) |
| 0.2 | 0.6 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.2 | 8.7 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.2 | 1.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 2.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 2.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 4.8 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.2 | 1.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 3.5 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.2 | 8.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.2 | 2.2 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 1.2 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.2 | 3.5 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 | 4.3 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.2 | 2.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 2.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 29.3 | GO:0006281 | DNA repair(GO:0006281) |
| 0.1 | 0.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 8.3 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 1.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) negative regulation of metaphase/anaphase transition of cell cycle(GO:1902100) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 2.0 | GO:0048641 | regulation of skeletal muscle tissue development(GO:0048641) |
| 0.1 | 10.4 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.1 | 3.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 7.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 3.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.9 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 3.0 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 2.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 5.9 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.1 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 4.1 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.1 | 0.9 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 12.3 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.1 | 3.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 1.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 2.0 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 1.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 1.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 13.4 | GO:0032259 | methylation(GO:0032259) |
| 0.1 | 2.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 1.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 1.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 4.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 15.8 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 1.8 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 1.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 1.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.0 | 4.0 | GO:0006974 | cellular response to DNA damage stimulus(GO:0006974) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.8 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 2.7 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 4.6 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 2.3 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 2.0 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 2.5 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 2.6 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 0.2 | GO:1901571 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) arachidonate transport(GO:1903963) |
| 0.0 | 2.1 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.0 | 0.7 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.2 | GO:0016441 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 1.9 | GO:0006417 | regulation of translation(GO:0006417) |
| 0.0 | 1.6 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.3 | GO:0045786 | negative regulation of cell cycle(GO:0045786) |
| 0.0 | 2.9 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 2.1 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 1.1 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 1.3 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 2.1 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 0.7 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 84.5 | GO:0042555 | MCM complex(GO:0042555) |
| 3.5 | 24.5 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 3.3 | 36.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 2.4 | 12.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 2.1 | 6.2 | GO:0098536 | deuterosome(GO:0098536) |
| 2.0 | 8.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.9 | 9.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 1.8 | 9.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.7 | 6.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.7 | 5.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 1.4 | 5.4 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 1.3 | 5.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 1.2 | 3.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 1.1 | 3.3 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 1.0 | 4.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.0 | 4.9 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.9 | 12.9 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.9 | 6.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.8 | 3.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.8 | 3.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.7 | 4.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.7 | 4.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.6 | 5.8 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.6 | 1.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.6 | 2.5 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.6 | 7.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.5 | 2.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.5 | 7.8 | GO:0070187 | telosome(GO:0070187) |
| 0.5 | 2.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 8.6 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.4 | 2.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.4 | 2.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 16.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.4 | 3.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 2.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 2.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 2.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 1.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 1.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.3 | 4.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.3 | 9.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 81.1 | GO:0000785 | chromatin(GO:0000785) |
| 0.2 | 3.3 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 4.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 1.5 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.2 | 1.1 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 2.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 2.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 54.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.2 | 3.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 3.5 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.2 | 20.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 29.7 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 1.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 3.4 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.1 | 3.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.5 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 2.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 13.8 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 0.5 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 6.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 7.4 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 68.6 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 1.3 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 4.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 189.4 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 7.0 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 24.2 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 4.2 | GO:0099568 | cell cortex(GO:0005938) cytoplasmic region(GO:0099568) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 43.9 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 4.4 | 109.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 3.0 | 20.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 2.7 | 10.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 2.4 | 7.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 2.2 | 6.7 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 2.0 | 8.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 1.7 | 8.3 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 1.7 | 19.8 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 1.6 | 4.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 1.6 | 4.7 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.6 | 20.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 1.5 | 4.6 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 1.5 | 11.9 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 1.3 | 5.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.3 | 6.7 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 1.3 | 7.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.2 | 4.8 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 1.1 | 3.4 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 1.1 | 4.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 1.1 | 7.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.8 | 2.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.8 | 3.2 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.8 | 3.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.8 | 3.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.7 | 3.7 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.7 | 3.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.7 | 6.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.7 | 4.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.6 | 1.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.6 | 1.9 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.6 | 1.3 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.6 | 8.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.6 | 6.3 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.6 | 9.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.5 | 25.2 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.5 | 1.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) efflux transmembrane transporter activity(GO:0015562) |
| 0.5 | 2.4 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.5 | 5.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.5 | 4.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.4 | 4.4 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.4 | 2.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 6.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 2.8 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.4 | 7.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.4 | 8.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.4 | 2.2 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.4 | 22.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.4 | 1.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.3 | 5.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 4.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.3 | 7.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.3 | 1.2 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.3 | 7.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.3 | 5.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.3 | 9.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 3.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 1.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 9.6 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.2 | 40.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.2 | 3.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 2.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 17.4 | GO:0016407 | acetyltransferase activity(GO:0016407) |
| 0.2 | 2.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 4.8 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 2.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 1.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.2 | 3.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.2 | 1.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 8.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.2 | 7.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 3.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 2.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 8.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 19.8 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.1 | 1.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 3.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 2.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 2.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 6.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 8.2 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.1 | 0.3 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.1 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 7.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 5.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 5.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 0.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 4.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 8.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.3 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 18.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.2 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.5 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.1 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 2.1 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 17.2 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 2.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 8.7 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 19.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 53.5 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 7.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.3 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 3.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 4.0 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.7 | GO:0019212 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.0 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 17.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.9 | 161.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.9 | 5.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.7 | 26.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.6 | 16.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.5 | 17.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.4 | 13.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 15.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.4 | 19.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.3 | 12.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 9.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.3 | 3.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.3 | 12.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 3.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 7.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 1.9 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 3.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 2.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 2.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 79.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 4.1 | 61.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 3.4 | 67.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 2.8 | 25.5 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 1.6 | 17.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.3 | 19.9 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.8 | 16.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.8 | 5.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.7 | 5.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.6 | 7.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.5 | 10.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 5.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.4 | 2.8 | REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | Genes involved in E2F mediated regulation of DNA replication |
| 0.4 | 6.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 2.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 5.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.3 | 4.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 3.5 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.2 | 6.6 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.2 | 1.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 2.5 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.2 | 5.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 2.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 12.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 3.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 4.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 5.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 6.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 2.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 4.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 4.4 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 1.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.7 | REACTOME RNA POL II TRANSCRIPTION | Genes involved in RNA Polymerase II Transcription |
| 0.1 | 1.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 2.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |