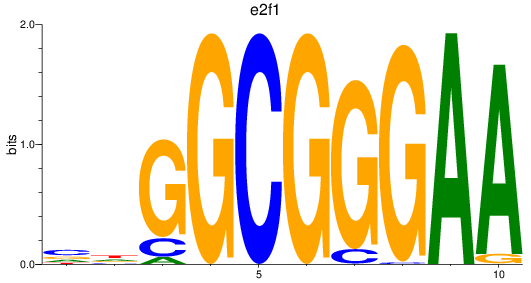
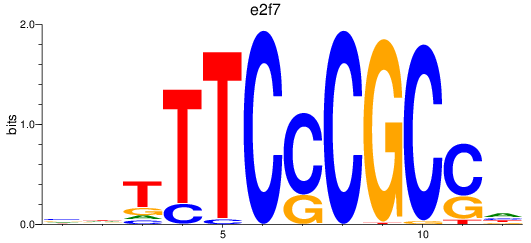
Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for e2f1_e2f7
Z-value: 3.46
Transcription factors associated with e2f1_e2f7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f1
|
ENSDARG00000103868 | E2F transcription factor 1 |
|
e2f7
|
ENSDARG00000008986 | E2F transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f7 | dr11_v1_chr4_-_2545310_2545345 | 0.79 | 5.5e-05 | Click! |
| e2f1 | dr11_v1_chr23_-_43486714_43486714 | 0.70 | 7.7e-04 | Click! |
Activity profile of e2f1_e2f7 motif
Sorted Z-values of e2f1_e2f7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_29498178 | 11.50 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr14_+_14841685 | 11.11 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr19_-_47570672 | 10.49 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr7_+_24814866 | 10.46 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr3_+_27027781 | 10.38 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr20_+_34770197 | 10.13 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr24_+_41690545 | 9.97 |
ENSDART00000160069
|
lama1
|
laminin, alpha 1 |
| chr7_+_24881680 | 9.60 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr19_+_627899 | 9.05 |
ENSDART00000148508
|
tert
|
telomerase reverse transcriptase |
| chr21_+_30721733 | 9.00 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr13_+_8255106 | 8.55 |
ENSDART00000080465
|
hells
|
helicase, lymphoid specific |
| chr8_+_15254564 | 8.52 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr6_+_12968101 | 8.30 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr8_-_18211605 | 7.95 |
ENSDART00000114177
|
CT025742.1
|
|
| chr18_-_16924221 | 7.90 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr6_-_33916756 | 7.49 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr13_-_86847 | 7.38 |
ENSDART00000158062
|
pole2
|
polymerase (DNA directed), epsilon 2 |
| chr18_+_17021391 | 7.38 |
ENSDART00000100160
|
chtf8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr5_-_67365333 | 7.35 |
ENSDART00000133438
|
unga
|
uracil DNA glycosylase a |
| chr1_-_38170997 | 6.98 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr24_-_35561672 | 6.96 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr11_+_1551603 | 6.85 |
ENSDART00000185383
ENSDART00000121489 ENSDART00000040577 |
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr20_+_13141408 | 6.84 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr13_+_48358467 | 6.73 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr3_+_25999477 | 6.69 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr20_+_33994580 | 6.58 |
ENSDART00000061729
|
si:dkey-97o5.1
|
si:dkey-97o5.1 |
| chr10_+_28428222 | 6.51 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr10_+_10728870 | 6.43 |
ENSDART00000109282
|
swi5
|
SWI5 homologous recombination repair protein |
| chr8_+_29635968 | 6.40 |
ENSDART00000139029
ENSDART00000091409 |
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr21_-_23017478 | 6.27 |
ENSDART00000024309
|
rb1
|
retinoblastoma 1 |
| chr3_-_35865040 | 6.19 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr2_+_32016516 | 6.11 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr13_-_25548733 | 5.85 |
ENSDART00000168099
ENSDART00000135788 ENSDART00000077655 |
mcmbp
|
minichromosome maintenance complex binding protein |
| chr20_-_32446406 | 5.82 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr13_+_26703922 | 5.76 |
ENSDART00000020946
|
fancl
|
Fanconi anemia, complementation group L |
| chr8_+_29636431 | 5.76 |
ENSDART00000133047
|
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr9_-_10068004 | 5.57 |
ENSDART00000011922
ENSDART00000162818 |
spopla
|
speckle-type POZ protein-like a |
| chr13_-_35908275 | 5.52 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr2_-_122154 | 5.46 |
ENSDART00000156248
ENSDART00000004071 |
znfl2a
|
zinc finger-like gene 2a |
| chr7_-_26518086 | 5.39 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr11_-_6970107 | 5.29 |
ENSDART00000171255
|
COMP
|
si:ch211-43f4.1 |
| chr3_+_43086548 | 5.28 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr17_-_11368662 | 5.22 |
ENSDART00000159061
ENSDART00000188694 ENSDART00000190932 |
si:ch211-185a18.2
|
si:ch211-185a18.2 |
| chr5_-_67365750 | 5.12 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr8_+_54013199 | 5.06 |
ENSDART00000158497
|
CABZ01079663.1
|
|
| chr20_-_36416922 | 4.95 |
ENSDART00000019145
|
lbr
|
lamin B receptor |
| chr14_-_22015232 | 4.92 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr22_-_21676364 | 4.89 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr22_-_20419660 | 4.80 |
ENSDART00000105520
|
pias4a
|
protein inhibitor of activated STAT, 4a |
| chr21_-_25213616 | 4.73 |
ENSDART00000122513
|
rfc2
|
replication factor C (activator 1) 2 |
| chr7_-_16582187 | 4.69 |
ENSDART00000131726
|
e2f8
|
E2F transcription factor 8 |
| chr13_-_39160018 | 4.67 |
ENSDART00000168795
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr3_+_13603272 | 4.61 |
ENSDART00000185084
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr18_+_17020967 | 4.56 |
ENSDART00000189168
|
chtf8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr9_+_56232548 | 4.51 |
ENSDART00000099276
|
cnot11
|
CCR4-NOT transcription complex, subunit 11 |
| chr4_+_13586455 | 4.51 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr6_-_51573975 | 4.50 |
ENSDART00000073865
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr13_-_39159810 | 4.47 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr20_-_38827623 | 4.45 |
ENSDART00000153310
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr15_+_17406920 | 4.42 |
ENSDART00000081059
|
rps6kb1b
|
ribosomal protein S6 kinase b, polypeptide 1b |
| chr22_-_22301672 | 4.28 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr5_+_39099380 | 4.24 |
ENSDART00000166657
|
bmp2k
|
BMP2 inducible kinase |
| chr7_+_27834130 | 4.20 |
ENSDART00000052656
|
rras2
|
RAS related 2 |
| chr3_-_49514874 | 4.17 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr7_+_71586485 | 4.14 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr23_+_17839187 | 4.11 |
ENSDART00000104647
|
prim1
|
DNA primase subunit 1 |
| chr3_-_61377127 | 4.10 |
ENSDART00000155932
|
si:dkey-111k8.2
|
si:dkey-111k8.2 |
| chr13_+_48359573 | 4.09 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr24_+_9693951 | 4.09 |
ENSDART00000082411
|
topbp1
|
DNA topoisomerase II binding protein 1 |
| chr24_-_13349464 | 4.08 |
ENSDART00000134482
ENSDART00000139212 |
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr19_-_2582858 | 4.00 |
ENSDART00000113829
|
cdca7b
|
cell division cycle associated 7b |
| chr6_-_39893501 | 3.97 |
ENSDART00000141611
ENSDART00000135631 ENSDART00000077662 ENSDART00000130613 |
myl6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr20_-_28698172 | 3.96 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr23_-_29357764 | 3.92 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr18_+_20226843 | 3.86 |
ENSDART00000100632
|
tle3a
|
transducin-like enhancer of split 3a |
| chr19_+_25465025 | 3.80 |
ENSDART00000018553
|
rpa3
|
replication protein A3 |
| chr24_+_27548474 | 3.79 |
ENSDART00000105774
ENSDART00000123368 |
ek1
|
eph-like kinase 1 |
| chr17_-_23412705 | 3.77 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr15_-_1484795 | 3.69 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr13_-_35907768 | 3.66 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr19_-_1947403 | 3.64 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr1_+_45740183 | 3.64 |
ENSDART00000149116
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr2_-_59285085 | 3.62 |
ENSDART00000131880
|
ftr34
|
finTRIM family, member 34 |
| chr18_-_30499489 | 3.62 |
ENSDART00000033746
|
gins2
|
GINS complex subunit 2 |
| chr8_+_52415603 | 3.59 |
ENSDART00000021604
ENSDART00000191424 |
gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr5_+_51248784 | 3.59 |
ENSDART00000159571
ENSDART00000189513 ENSDART00000092285 |
msh3
|
mutS homolog 3 (E. coli) |
| chr19_+_1873059 | 3.57 |
ENSDART00000145246
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr6_-_53334259 | 3.53 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr16_+_37891735 | 3.51 |
ENSDART00000178753
ENSDART00000142104 |
cep162
|
centrosomal protein 162 |
| chr7_+_55518519 | 3.51 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr20_-_37933237 | 3.50 |
ENSDART00000142567
ENSDART00000036371 ENSDART00000061445 |
angel2
|
angel homolog 2 (Drosophila) |
| chr23_+_29358188 | 3.48 |
ENSDART00000189242
|
tardbpl
|
TAR DNA binding protein, like |
| chr18_-_8380090 | 3.47 |
ENSDART00000141581
ENSDART00000081143 |
sephs1
|
selenophosphate synthetase 1 |
| chr16_-_4770233 | 3.46 |
ENSDART00000193228
|
rpa2
|
replication protein A2 |
| chr15_-_1485086 | 3.46 |
ENSDART00000191651
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr1_+_14020445 | 3.40 |
ENSDART00000079716
|
hpf1
|
histone PARylation factor 1 |
| chr4_-_2380173 | 3.38 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr16_-_4769877 | 3.36 |
ENSDART00000149421
ENSDART00000054078 |
rpa2
|
replication protein A2 |
| chr23_+_25428513 | 3.35 |
ENSDART00000144554
|
fmnl3
|
formin-like 3 |
| chr5_+_69697800 | 3.35 |
ENSDART00000178736
ENSDART00000162519 |
znf1005
|
zinc finger protein 1005 |
| chr8_+_28593707 | 3.31 |
ENSDART00000097213
|
tcf15
|
transcription factor 15 |
| chr24_-_13349802 | 3.29 |
ENSDART00000164729
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr5_+_41477526 | 3.28 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr12_+_17603528 | 3.26 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr21_-_27413294 | 3.25 |
ENSDART00000131646
|
slc29a2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr16_-_41990421 | 3.23 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr4_+_13586689 | 3.22 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr22_+_1911269 | 3.20 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr8_+_28547687 | 3.18 |
ENSDART00000110291
|
srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr5_-_7712160 | 3.15 |
ENSDART00000168820
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
| chr6_+_33931740 | 3.14 |
ENSDART00000130492
ENSDART00000151213 |
orc1
|
origin recognition complex, subunit 1 |
| chr7_+_1550966 | 3.12 |
ENSDART00000177863
ENSDART00000126840 |
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr19_+_1872794 | 3.07 |
ENSDART00000013217
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr25_-_19585010 | 3.07 |
ENSDART00000021340
|
sycp3
|
synaptonemal complex protein 3 |
| chr8_+_18615938 | 3.06 |
ENSDART00000089161
|
si:ch211-232d19.4
|
si:ch211-232d19.4 |
| chr24_+_26329018 | 3.05 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr3_+_34988670 | 3.04 |
ENSDART00000011319
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr22_-_5655680 | 3.03 |
ENSDART00000159629
|
mcm2
|
minichromosome maintenance complex component 2 |
| chr9_-_33081978 | 3.02 |
ENSDART00000100918
|
zgc:172053
|
zgc:172053 |
| chr4_-_64123545 | 2.94 |
ENSDART00000170040
|
BX914205.2
|
|
| chr5_-_67365006 | 2.92 |
ENSDART00000136116
|
unga
|
uracil DNA glycosylase a |
| chr19_+_48024457 | 2.88 |
ENSDART00000163823
|
kpnb1
|
karyopherin (importin) beta 1 |
| chr23_-_3758637 | 2.84 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr4_+_38344 | 2.84 |
ENSDART00000170197
ENSDART00000175348 |
phtf2
|
putative homeodomain transcription factor 2 |
| chr16_+_4770266 | 2.83 |
ENSDART00000038036
|
ube3d
|
ubiquitin protein ligase E3D |
| chr17_+_25833947 | 2.82 |
ENSDART00000044328
ENSDART00000154604 |
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr2_+_32016256 | 2.82 |
ENSDART00000005143
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr19_-_5358443 | 2.81 |
ENSDART00000105036
|
cyt1l
|
type I cytokeratin, enveloping layer, like |
| chr9_-_746317 | 2.79 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr19_-_20199167 | 2.79 |
ENSDART00000155527
|
si:ch211-155k24.9
|
si:ch211-155k24.9 |
| chr7_+_41812636 | 2.77 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr15_-_26538989 | 2.75 |
ENSDART00000032880
|
rpa1
|
replication protein A1 |
| chr22_-_15704704 | 2.75 |
ENSDART00000017838
ENSDART00000130238 |
safb
|
scaffold attachment factor B |
| chr1_+_58613972 | 2.74 |
ENSDART00000162460
|
si:ch73-221f6.3
|
si:ch73-221f6.3 |
| chr1_+_11822 | 2.73 |
ENSDART00000166393
|
cep97
|
centrosomal protein 97 |
| chr3_-_16719244 | 2.72 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr8_+_10304981 | 2.72 |
ENSDART00000160766
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr1_-_44000940 | 2.70 |
ENSDART00000134932
ENSDART00000138447 |
zgc:66472
|
zgc:66472 |
| chr17_-_15189397 | 2.68 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr14_-_16775158 | 2.67 |
ENSDART00000113711
ENSDART00000144781 ENSDART00000160411 |
mrnip
|
MRN complex interacting protein |
| chr25_+_8447565 | 2.67 |
ENSDART00000142090
|
fanci
|
Fanconi anemia, complementation group I |
| chr13_+_42602406 | 2.66 |
ENSDART00000133388
ENSDART00000147996 |
mlh1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) |
| chr13_+_28785814 | 2.66 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr5_-_19400166 | 2.64 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr18_-_7456378 | 2.63 |
ENSDART00000081459
|
pdp2
|
putative pyruvate dehydrogenase phosphatase isoenzyme 2 |
| chr5_+_34549365 | 2.61 |
ENSDART00000009500
|
aif1l
|
allograft inflammatory factor 1-like |
| chr9_-_54361357 | 2.61 |
ENSDART00000149813
|
rad50
|
RAD50 homolog, double strand break repair protein |
| chr10_+_21434649 | 2.60 |
ENSDART00000193938
ENSDART00000064558 |
etf1b
|
eukaryotic translation termination factor 1b |
| chr7_+_41812817 | 2.59 |
ENSDART00000174165
|
orc6
|
origin recognition complex, subunit 6 |
| chr1_+_24557414 | 2.53 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr2_+_25378457 | 2.51 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr1_+_11992 | 2.51 |
ENSDART00000161842
|
cep97
|
centrosomal protein 97 |
| chr23_+_4483083 | 2.50 |
ENSDART00000092389
|
nup210
|
nucleoporin 210 |
| chr18_+_30421528 | 2.50 |
ENSDART00000140908
|
gse1
|
Gse1 coiled-coil protein |
| chr2_-_59303338 | 2.47 |
ENSDART00000100987
ENSDART00000140840 |
ftr35
|
finTRIM family, member 35 |
| chr9_-_30555725 | 2.46 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr12_+_46883785 | 2.45 |
ENSDART00000008312
|
fam53b
|
family with sequence similarity 53, member B |
| chr21_+_5862476 | 2.44 |
ENSDART00000065848
|
wdr38
|
WD repeat domain 38 |
| chr23_-_6722101 | 2.44 |
ENSDART00000157828
|
baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr25_-_12824656 | 2.44 |
ENSDART00000171801
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr13_+_6188759 | 2.43 |
ENSDART00000161062
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr3_-_5964557 | 2.41 |
ENSDART00000184738
|
BX284638.1
|
|
| chr13_+_111212 | 2.39 |
ENSDART00000167840
|
dnaaf2
|
dynein, axonemal, assembly factor 2 |
| chr10_+_2799285 | 2.39 |
ENSDART00000030709
|
pnx
|
posterior neuron-specific homeobox |
| chr6_+_22326624 | 2.34 |
ENSDART00000020333
|
rae1
|
ribonucleic acid export 1 |
| chr11_-_3308569 | 2.33 |
ENSDART00000036581
|
cdk2
|
cyclin-dependent kinase 2 |
| chr11_+_45153104 | 2.31 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr22_-_4439311 | 2.28 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr25_+_35020529 | 2.28 |
ENSDART00000158016
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr11_+_13176568 | 2.26 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr5_+_24047292 | 2.25 |
ENSDART00000029889
|
ctdnep1a
|
CTD nuclear envelope phosphatase 1a |
| chr8_-_48847772 | 2.25 |
ENSDART00000122458
|
wrap73
|
WD repeat containing, antisense to TP73 |
| chr1_+_49415281 | 2.24 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr9_-_2892250 | 2.24 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr5_+_34549845 | 2.21 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr5_-_23179319 | 2.21 |
ENSDART00000161883
ENSDART00000136260 |
si:dkey-114c15.5
|
si:dkey-114c15.5 |
| chr24_+_26328787 | 2.21 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr16_+_55078477 | 2.21 |
ENSDART00000170074
|
azi2
|
5-azacytidine induced 2 |
| chr3_-_7897305 | 2.18 |
ENSDART00000169757
|
ubn2b
|
ubinuclein 2b |
| chr20_-_54245256 | 2.17 |
ENSDART00000170482
|
prpf39
|
PRP39 pre-mRNA processing factor 39 homolog (yeast) |
| chr22_-_22301929 | 2.15 |
ENSDART00000142027
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr20_-_33487729 | 2.15 |
ENSDART00000061843
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr16_+_43139504 | 2.14 |
ENSDART00000065643
|
dbf4
|
DBF4 zinc finger |
| chr7_-_37919475 | 2.14 |
ENSDART00000052368
|
heatr3
|
HEAT repeat containing 3 |
| chr6_+_18544791 | 2.14 |
ENSDART00000167463
ENSDART00000169599 |
atad5b
|
ATPase family, AAA domain containing 5b |
| chr8_+_49766338 | 2.09 |
ENSDART00000060657
|
rmi1
|
RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) |
| chr6_-_6976096 | 2.09 |
ENSDART00000151822
ENSDART00000039443 ENSDART00000177960 |
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr16_-_4836218 | 2.08 |
ENSDART00000054077
|
tpbgb
|
trophoblast glycoprotein b |
| chr7_+_41812190 | 2.07 |
ENSDART00000113732
ENSDART00000174137 |
orc6
|
origin recognition complex, subunit 6 |
| chr20_+_46427984 | 2.06 |
ENSDART00000060706
ENSDART00000143858 |
rad51
|
RAD51 recombinase |
| chr4_-_2545310 | 2.05 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr11_-_27702778 | 2.01 |
ENSDART00000045942
ENSDART00000125352 |
phf2
|
PHD finger protein 2 |
| chr17_+_41992054 | 2.00 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr16_-_29557338 | 2.00 |
ENSDART00000058888
|
hormad1
|
HORMA domain containing 1 |
| chr6_-_9282080 | 1.99 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr25_+_35062353 | 1.98 |
ENSDART00000089844
|
zgc:113983
|
zgc:113983 |
| chr6_-_33931696 | 1.97 |
ENSDART00000057290
|
prpf38a
|
pre-mRNA processing factor 38A |
| chr22_+_2511045 | 1.95 |
ENSDART00000106425
|
zgc:173726
|
zgc:173726 |
| chr3_-_18737126 | 1.93 |
ENSDART00000055767
|
e4f1
|
E4F transcription factor 1 |
| chr3_+_60589157 | 1.93 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f1_e2f7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0006290 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 3.5 | 10.4 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 3.2 | 35.1 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 3.1 | 15.4 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 3.0 | 9.1 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 2.9 | 11.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 2.5 | 2.5 | GO:0009219 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) pyrimidine deoxyribonucleotide metabolic process(GO:0009219) |
| 2.0 | 22.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 1.8 | 5.4 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 1.8 | 9.0 | GO:0000012 | single strand break repair(GO:0000012) |
| 1.6 | 11.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 1.5 | 10.8 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 1.5 | 4.5 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 1.4 | 10.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 1.4 | 13.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 1.3 | 6.7 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 1.2 | 1.2 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 1.1 | 2.1 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 1.0 | 7.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 1.0 | 4.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.0 | 12.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 1.0 | 4.0 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 1.0 | 4.8 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.9 | 2.7 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.8 | 17.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.8 | 7.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.8 | 2.3 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.7 | 3.4 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.7 | 2.0 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.7 | 2.6 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.6 | 13.6 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.6 | 3.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.6 | 1.7 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.6 | 1.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.5 | 3.2 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.5 | 3.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.5 | 2.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.5 | 2.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.5 | 10.8 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.5 | 2.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.5 | 2.0 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.5 | 1.4 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.5 | 2.8 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.5 | 2.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.5 | 2.3 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.4 | 2.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.4 | 1.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.4 | 3.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.4 | 3.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.4 | 1.2 | GO:0045141 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.4 | 1.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.4 | 10.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.4 | 1.6 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.4 | 3.9 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.4 | 1.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 1.1 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.4 | 2.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 3.5 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.3 | 1.7 | GO:0051591 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.3 | 1.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.3 | 1.6 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.3 | 3.8 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.3 | 2.8 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.3 | 3.0 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.3 | 39.5 | GO:0006260 | DNA replication(GO:0006260) |
| 0.3 | 1.5 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.3 | 5.6 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.3 | 1.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.3 | 1.4 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.3 | 16.1 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.3 | 1.6 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.3 | 8.6 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.3 | 1.8 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.3 | 0.5 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.3 | 1.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.3 | 2.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 8.0 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.2 | 4.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 4.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.2 | 1.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 2.7 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.2 | 6.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 5.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.2 | 1.3 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 8.6 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.2 | 12.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.2 | 5.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 1.3 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.2 | 4.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 5.0 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 2.3 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.1 | 1.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 2.4 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 4.1 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 1.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 2.7 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 3.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 1.4 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.1 | 2.3 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.1 | 0.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 1.6 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 5.5 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.1 | 2.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 3.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 1.0 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.4 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 4.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 2.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 1.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 1.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 5.1 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.1 | 4.4 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.1 | 2.2 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 1.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 2.2 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.8 | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902253) |
| 0.1 | 0.6 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 4.5 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 3.2 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.1 | 0.2 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 | 0.9 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.2 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 3.3 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 0.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 4.4 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.0 | 0.5 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.9 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.6 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.9 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 5.7 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 5.5 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 1.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 3.1 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.6 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 1.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 3.5 | GO:0016458 | gene silencing(GO:0016458) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.8 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 2.0 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 2.7 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 6.3 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 1.6 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 1.1 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 3.2 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 3.9 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 5.3 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 2.9 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 1.9 | GO:1903507 | negative regulation of transcription, DNA-templated(GO:0045892) negative regulation of RNA biosynthetic process(GO:1902679) negative regulation of nucleic acid-templated transcription(GO:1903507) |
| 0.0 | 0.3 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 3.0 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 1.9 | GO:0040008 | regulation of growth(GO:0040008) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.7 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 4.0 | 11.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 3.0 | 9.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 2.7 | 13.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 2.3 | 6.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 2.3 | 41.0 | GO:0042555 | MCM complex(GO:0042555) |
| 2.2 | 8.9 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 2.1 | 8.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.4 | 7.2 | GO:0000811 | GINS complex(GO:0000811) |
| 1.4 | 4.1 | GO:1990077 | primosome complex(GO:1990077) |
| 1.4 | 9.5 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 1.1 | 8.0 | GO:0035101 | FACT complex(GO:0035101) |
| 1.0 | 7.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 1.0 | 11.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.9 | 6.6 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.9 | 5.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.8 | 3.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.8 | 4.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.8 | 6.4 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.7 | 3.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.7 | 4.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.6 | 6.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.6 | 2.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.5 | 2.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.5 | 2.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.5 | 12.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.5 | 6.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.5 | 3.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.5 | 2.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 1.3 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.4 | 9.4 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.3 | 2.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.3 | 3.0 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.3 | 1.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.3 | 1.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 1.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 2.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 2.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 4.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 2.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 2.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.2 | 3.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 5.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 1.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 1.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 5.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 3.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.7 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.1 | 1.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 15.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 8.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 10.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 4.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 10.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 3.9 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 3.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 2.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.0 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 1.0 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 2.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 5.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 2.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 2.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 8.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 6.1 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 24.0 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 1.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 123.6 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.8 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 17.3 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 5.2 | GO:0019866 | organelle inner membrane(GO:0019866) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.4 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 4.2 | 12.7 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 3.6 | 3.6 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 3.1 | 22.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 3.0 | 9.1 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 2.5 | 7.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 1.7 | 6.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 1.7 | 8.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 1.7 | 8.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 1.6 | 9.7 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 1.5 | 4.5 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 1.2 | 21.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.9 | 2.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.8 | 5.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.8 | 6.7 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.8 | 10.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.7 | 2.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.7 | 2.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.6 | 8.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.6 | 2.4 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.6 | 5.4 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.6 | 12.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.5 | 5.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.5 | 2.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.5 | 4.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.5 | 4.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 3.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 18.3 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.4 | 1.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.4 | 7.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.4 | 1.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.4 | 2.5 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.4 | 12.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.4 | 4.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.4 | 4.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.3 | 9.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 3.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 3.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 2.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.3 | 1.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 1.8 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.3 | 2.6 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 1.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.3 | 3.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 4.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.3 | 2.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 2.7 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.2 | 1.9 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.2 | 3.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.2 | 3.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 3.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 1.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 8.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.2 | 1.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 5.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 9.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 1.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 7.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 1.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 4.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 7.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 13.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.2 | 5.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 3.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 8.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 3.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.6 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 1.3 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.1 | 17.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 10.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 2.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 11.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 4.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 5.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 4.6 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 23.9 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 1.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 4.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 2.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 2.8 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 6.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 10.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.3 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 1.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.9 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 3.4 | GO:0003779 | actin binding(GO:0003779) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 10.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.7 | 59.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.7 | 7.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 25.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.4 | 3.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 17.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.3 | 4.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 5.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 3.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 1.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 2.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 6.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 2.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 11.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 7.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 4.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 2.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 6.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 42.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.7 | 20.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.3 | 12.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 1.2 | 28.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.0 | 4.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 1.0 | 13.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 1.0 | 11.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 1.0 | 9.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.9 | 6.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.7 | 7.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.6 | 3.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.6 | 2.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.6 | 2.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 5.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.4 | 6.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.4 | 6.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.4 | 4.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 3.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 5.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 4.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 4.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 2.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 1.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 11.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 1.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 2.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 8.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 5.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |