Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
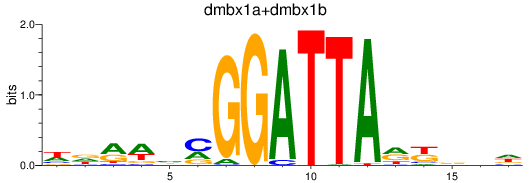
Results for dmbx1a+dmbx1b
Z-value: 0.54
Transcription factors associated with dmbx1a+dmbx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dmbx1b
|
ENSDARG00000002510 | diencephalon/mesencephalon homeobox 1b |
|
dmbx1a
|
ENSDARG00000009922 | diencephalon/mesencephalon homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dmbx1b | dr11_v1_chr6_-_3573716_3573716 | 0.86 | 1.8e-06 | Click! |
| dmbx1a | dr11_v1_chr2_-_10188598_10188598 | -0.49 | 3.3e-02 | Click! |
Activity profile of dmbx1a+dmbx1b motif
Sorted Z-values of dmbx1a+dmbx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_1805421 | 3.82 |
ENSDART00000173143
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr14_-_46238186 | 1.15 |
ENSDART00000173245
|
si:ch211-113d11.6
|
si:ch211-113d11.6 |
| chr17_+_17955063 | 1.04 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr20_-_33515890 | 0.87 |
ENSDART00000159987
|
si:dkey-65b13.13
|
si:dkey-65b13.13 |
| chr1_-_45920632 | 0.78 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr1_-_45177373 | 0.73 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr14_+_2487672 | 0.64 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr2_+_51783120 | 0.63 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr21_-_3770636 | 0.59 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr1_-_12126535 | 0.57 |
ENSDART00000164817
ENSDART00000015251 |
mttp
|
microsomal triglyceride transfer protein |
| chr15_-_8191841 | 0.57 |
ENSDART00000156663
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr12_+_3848109 | 0.55 |
ENSDART00000144067
ENSDART00000192042 |
cabp5a
|
calcium binding protein 5a |
| chr5_-_5243079 | 0.53 |
ENSDART00000130576
ENSDART00000164377 |
mvb12ba
|
multivesicular body subunit 12Ba |
| chr2_+_243778 | 0.51 |
ENSDART00000182262
|
CABZ01085887.1
|
|
| chr6_-_10320676 | 0.51 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr7_+_19903924 | 0.50 |
ENSDART00000159112
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr6_-_39653972 | 0.49 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr5_+_69808763 | 0.47 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr14_+_11909966 | 0.44 |
ENSDART00000171829
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr3_+_32142382 | 0.42 |
ENSDART00000133035
|
syt5a
|
synaptotagmin Va |
| chr3_+_16762483 | 0.40 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr15_-_8191992 | 0.40 |
ENSDART00000155381
ENSDART00000190714 |
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr5_-_69180587 | 0.37 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr20_-_1141722 | 0.36 |
ENSDART00000152675
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr25_-_19068557 | 0.36 |
ENSDART00000184780
|
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr3_+_15296824 | 0.35 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr4_-_42039794 | 0.35 |
ENSDART00000110313
|
si:dkeyp-85d8.5
|
si:dkeyp-85d8.5 |
| chr23_-_26652273 | 0.35 |
ENSDART00000112124
ENSDART00000111029 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr8_-_30424182 | 0.34 |
ENSDART00000099021
|
dock8
|
dedicator of cytokinesis 8 |
| chr12_+_31729498 | 0.33 |
ENSDART00000188546
ENSDART00000182562 ENSDART00000186147 |
RNF157
|
si:dkey-49c17.3 |
| chr11_+_27973055 | 0.32 |
ENSDART00000135135
|
alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr12_+_31729075 | 0.31 |
ENSDART00000152973
|
RNF157
|
si:dkey-49c17.3 |
| chr23_+_45512825 | 0.31 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
| chr1_+_52560549 | 0.31 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr13_-_14269626 | 0.30 |
ENSDART00000079176
|
gfra4a
|
GDNF family receptor alpha 4a |
| chr11_-_3366782 | 0.30 |
ENSDART00000127982
|
suox
|
sulfite oxidase |
| chr10_-_18468385 | 0.29 |
ENSDART00000183302
|
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr1_+_47585700 | 0.29 |
ENSDART00000153746
ENSDART00000084457 |
sh3pxd2aa
|
SH3 and PX domains 2Aa |
| chr17_+_10593398 | 0.28 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr8_+_54263946 | 0.28 |
ENSDART00000193119
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr13_+_1015749 | 0.27 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr18_-_10713230 | 0.27 |
ENSDART00000183646
|
rbm28
|
RNA binding motif protein 28 |
| chr18_+_5172848 | 0.26 |
ENSDART00000190642
|
CABZ01080599.1
|
|
| chr7_+_19904136 | 0.26 |
ENSDART00000173452
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr22_+_6740039 | 0.25 |
ENSDART00000144122
|
CT583625.2
|
|
| chr16_+_48678655 | 0.24 |
ENSDART00000150156
|
tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr7_+_56577906 | 0.23 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr21_-_20342096 | 0.23 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr13_+_40815012 | 0.22 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr15_-_7337148 | 0.22 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr10_-_37075361 | 0.22 |
ENSDART00000132023
|
myo18aa
|
myosin XVIIIAa |
| chr5_-_30481263 | 0.22 |
ENSDART00000086734
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr4_-_49133107 | 0.22 |
ENSDART00000150806
|
znf1146
|
zinc finger protein 1146 |
| chr18_+_21951551 | 0.22 |
ENSDART00000146261
|
ranbp10
|
RAN binding protein 10 |
| chr10_+_24504292 | 0.20 |
ENSDART00000090059
|
mtus2a
|
microtubule associated tumor suppressor candidate 2a |
| chr18_-_10713414 | 0.20 |
ENSDART00000034817
|
rbm28
|
RNA binding motif protein 28 |
| chr5_-_69180227 | 0.20 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr3_-_41995321 | 0.19 |
ENSDART00000192277
|
ttyh3a
|
tweety family member 3a |
| chr13_+_50151407 | 0.18 |
ENSDART00000031858
|
gpr137ba
|
G protein-coupled receptor 137Ba |
| chr15_+_45640906 | 0.17 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr15_-_41734639 | 0.17 |
ENSDART00000154230
ENSDART00000167443 |
ftr90
|
finTRIM family, member 90 |
| chr18_+_45781115 | 0.16 |
ENSDART00000151699
ENSDART00000179887 |
rpl35a
|
ribosomal protein L35a |
| chr19_+_3140313 | 0.16 |
ENSDART00000125504
|
zgc:86598
|
zgc:86598 |
| chr7_+_18989241 | 0.15 |
ENSDART00000053213
|
zgc:194252
|
zgc:194252 |
| chr17_-_21617618 | 0.15 |
ENSDART00000109031
|
gpr26
|
G protein-coupled receptor 26 |
| chr18_+_15644559 | 0.14 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr9_-_38587275 | 0.13 |
ENSDART00000077446
|
si:dkey-101k6.5
|
si:dkey-101k6.5 |
| chr7_+_65121743 | 0.13 |
ENSDART00000108471
|
ipo7
|
importin 7 |
| chr7_-_21905851 | 0.13 |
ENSDART00000111066
ENSDART00000020288 |
epoa
|
erythropoietin a |
| chr12_+_33894396 | 0.13 |
ENSDART00000130853
ENSDART00000152988 |
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr20_+_12830448 | 0.12 |
ENSDART00000164754
|
BX547930.4
|
|
| chr16_-_12236362 | 0.12 |
ENSDART00000114759
|
lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr3_+_19914332 | 0.12 |
ENSDART00000078982
|
vat1
|
vesicle amine transport 1 |
| chr7_+_19483277 | 0.11 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr7_+_38395197 | 0.11 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr7_-_72423666 | 0.10 |
ENSDART00000191214
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr12_+_292452 | 0.09 |
ENSDART00000189944
|
CABZ01058222.1
|
|
| chr5_-_22602979 | 0.09 |
ENSDART00000146287
|
nono
|
non-POU domain containing, octamer-binding |
| chr11_-_29082429 | 0.09 |
ENSDART00000041443
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr23_-_27050083 | 0.09 |
ENSDART00000142324
ENSDART00000133249 ENSDART00000138751 ENSDART00000128718 |
zgc:66440
|
zgc:66440 |
| chr20_-_31308805 | 0.08 |
ENSDART00000147045
|
hpcal1
|
hippocalcin-like 1 |
| chr11_-_29082175 | 0.08 |
ENSDART00000123245
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr10_+_24504464 | 0.08 |
ENSDART00000141387
|
mtus2a
|
microtubule associated tumor suppressor candidate 2a |
| chr21_-_20341836 | 0.08 |
ENSDART00000176689
|
rbp4l
|
retinol binding protein 4, like |
| chr7_-_54320088 | 0.08 |
ENSDART00000172396
|
fadd
|
Fas (tnfrsf6)-associated via death domain |
| chr21_-_28340977 | 0.07 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr18_+_15182965 | 0.07 |
ENSDART00000137018
|
si:ch73-62b13.1
|
si:ch73-62b13.1 |
| chr19_+_24324967 | 0.07 |
ENSDART00000090081
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr25_+_11456696 | 0.07 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr22_+_2709467 | 0.07 |
ENSDART00000141416
|
znf1171
|
zinc finger protein 1171 |
| chr20_-_31067306 | 0.06 |
ENSDART00000014163
|
fndc1
|
fibronectin type III domain containing 1 |
| chr5_-_25733745 | 0.06 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr16_+_16265850 | 0.06 |
ENSDART00000181265
|
setd2
|
SET domain containing 2 |
| chr5_-_8817841 | 0.06 |
ENSDART00000170921
|
fgf10b
|
fibroblast growth factor 10b |
| chr17_-_31058900 | 0.05 |
ENSDART00000134998
ENSDART00000104307 ENSDART00000172721 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr1_+_35949269 | 0.05 |
ENSDART00000180628
|
BX276105.3
|
|
| chr11_+_29671661 | 0.05 |
ENSDART00000024318
ENSDART00000165024 |
rnf207a
|
ring finger protein 207a |
| chr4_+_76671012 | 0.05 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr4_+_47385355 | 0.05 |
ENSDART00000163413
|
znf1122
|
zinc finger protein 1122 |
| chr4_-_59152337 | 0.04 |
ENSDART00000156770
|
znf1149
|
zinc finger protein 1149 |
| chr5_+_25733774 | 0.04 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr3_-_21402279 | 0.04 |
ENSDART00000164513
|
CT573446.1
|
|
| chr7_-_35409027 | 0.04 |
ENSDART00000128334
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr13_+_1726953 | 0.04 |
ENSDART00000103004
|
zmp:0000000760
|
zmp:0000000760 |
| chr17_-_8656155 | 0.04 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr3_+_34919810 | 0.02 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr20_-_9963713 | 0.02 |
ENSDART00000104234
|
gjd2b
|
gap junction protein delta 2b |
| chr19_+_2631565 | 0.02 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr25_-_32381642 | 0.01 |
ENSDART00000133872
ENSDART00000006124 |
cops2
|
COP9 signalosome subunit 2 |
| chr2_+_44972720 | 0.01 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr4_+_33012407 | 0.01 |
ENSDART00000151873
|
si:dkey-26h11.2
|
si:dkey-26h11.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dmbx1a+dmbx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.5 | GO:0044803 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.1 | 0.9 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.4 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.3 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.6 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.2 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.3 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 3.7 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.2 | GO:0002029 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.3 | GO:0006801 | superoxide metabolic process(GO:0006801) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.4 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.2 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.0 | 0.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |