Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for dlx2a+dlx2b_dlx4a+dlx4b+dlx6a
Z-value: 0.92
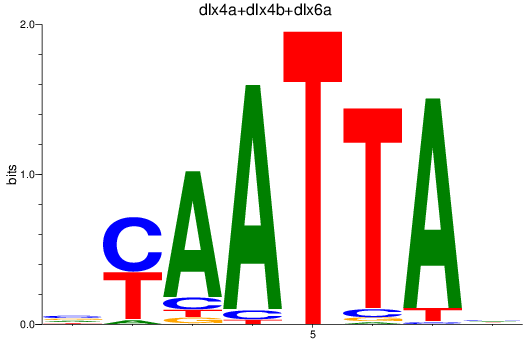
Transcription factors associated with dlx2a+dlx2b_dlx4a+dlx4b+dlx6a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx2b
|
ENSDARG00000017174 | distal-less homeobox 2b |
|
dlx2a
|
ENSDARG00000079964 | distal-less homeobox 2a |
|
dlx2b
|
ENSDARG00000116017 | distal-less homeobox 2b |
|
dlx4a
|
ENSDARG00000011956 | distal-less homeobox 4a |
|
dlx6a
|
ENSDARG00000042291 | distal-less homeobox 6a |
|
dlx4b
|
ENSDARG00000071560 | distal-less homeobox 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx4a | dr11_v1_chr3_+_34670076_34670076 | 0.75 | 2.3e-04 | Click! |
| dlx6a | dr11_v1_chr19_+_41464870_41464870 | 0.61 | 5.2e-03 | Click! |
| dlx4b | dr11_v1_chr12_-_5728755_5728755 | -0.32 | 1.8e-01 | Click! |
| dlx2a | dr11_v1_chr9_-_3400727_3400729 | -0.31 | 2.0e-01 | Click! |
| dlx2b | dr11_v1_chr1_-_30979707_30979707 | 0.01 | 9.5e-01 | Click! |
Activity profile of dlx2a+dlx2b_dlx4a+dlx4b+dlx6a motif
Sorted Z-values of dlx2a+dlx2b_dlx4a+dlx4b+dlx6a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_30244356 | 5.48 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr25_+_29160102 | 4.15 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr1_-_43905252 | 4.06 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr16_+_5774977 | 3.65 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr20_-_40755614 | 3.53 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr13_+_25449681 | 3.24 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr19_+_10339538 | 2.85 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr23_-_38497705 | 2.67 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr24_-_24849091 | 2.63 |
ENSDART00000133649
ENSDART00000038290 |
crhb
|
corticotropin releasing hormone b |
| chr12_+_24342303 | 2.56 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr10_+_29698467 | 2.41 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr2_-_33645411 | 2.31 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr1_-_50859053 | 2.16 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr2_+_37227011 | 2.15 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr22_+_20427170 | 2.15 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr15_-_6247775 | 2.13 |
ENSDART00000148350
|
dscamb
|
Down syndrome cell adhesion molecule b |
| chr7_-_28148310 | 2.06 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr16_+_31804590 | 2.03 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr14_-_30587814 | 2.01 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr13_-_40401870 | 1.93 |
ENSDART00000128951
|
nkx3.3
|
NK3 homeobox 3 |
| chr6_-_41229787 | 1.88 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr20_-_29420713 | 1.87 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr13_+_38430466 | 1.85 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr20_-_9462433 | 1.83 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr8_-_46894362 | 1.62 |
ENSDART00000111124
|
acot7
|
acyl-CoA thioesterase 7 |
| chr23_+_28731379 | 1.62 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr16_+_13818500 | 1.56 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr4_+_12612723 | 1.52 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr16_-_13921589 | 1.45 |
ENSDART00000023543
|
rcvrn2
|
recoverin 2 |
| chr16_+_17389116 | 1.45 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr4_+_7391110 | 1.44 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr7_+_30787903 | 1.42 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr19_-_5103141 | 1.39 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr17_-_12389259 | 1.38 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr16_-_47483142 | 1.32 |
ENSDART00000147072
|
cthrc1b
|
collagen triple helix repeat containing 1b |
| chr2_+_50608099 | 1.32 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr17_-_16965809 | 1.32 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr10_-_34871737 | 1.31 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr3_-_32337653 | 1.29 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr20_+_30445971 | 1.27 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr19_-_7358184 | 1.26 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr1_+_16397063 | 1.23 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr23_-_29824146 | 1.22 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr1_+_52929185 | 1.22 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr20_-_28800999 | 1.20 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr19_-_32804535 | 1.19 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr4_+_11384891 | 1.15 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr1_+_16396870 | 1.12 |
ENSDART00000189245
ENSDART00000113266 |
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr1_-_5455498 | 1.11 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr15_+_9053059 | 1.11 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr18_+_15644559 | 1.07 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr10_+_3049636 | 1.07 |
ENSDART00000081794
ENSDART00000183167 ENSDART00000191634 ENSDART00000183514 |
rasgrf2a
|
Ras protein-specific guanine nucleotide-releasing factor 2a |
| chr14_-_17622080 | 1.05 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr18_+_9637744 | 1.04 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr6_-_14292307 | 1.03 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr15_-_34408777 | 1.03 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr2_-_15324837 | 1.02 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr10_+_41765616 | 0.99 |
ENSDART00000170682
|
rnf34b
|
ring finger protein 34b |
| chr20_-_32270866 | 0.96 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr21_+_28958471 | 0.93 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr8_-_20230559 | 0.91 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr21_-_42100471 | 0.90 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr1_-_44701313 | 0.89 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr6_+_36807861 | 0.88 |
ENSDART00000161708
|
si:ch73-29l19.1
|
si:ch73-29l19.1 |
| chr2_-_16380283 | 0.87 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr23_+_20689255 | 0.87 |
ENSDART00000182420
|
usp21
|
ubiquitin specific peptidase 21 |
| chr4_+_26629368 | 0.86 |
ENSDART00000183575
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr3_+_33341640 | 0.86 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr23_+_20563779 | 0.86 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr17_-_20717845 | 0.85 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr3_+_34919810 | 0.85 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr19_-_205104 | 0.85 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr20_-_45060241 | 0.84 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr1_-_22512063 | 0.83 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr22_-_11136625 | 0.81 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr1_+_11977426 | 0.81 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr16_+_20161805 | 0.80 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr2_-_23004286 | 0.79 |
ENSDART00000134664
ENSDART00000110373 ENSDART00000185833 ENSDART00000187235 |
znf414
mllt1b
|
zinc finger protein 414 MLLT1, super elongation complex subunit b |
| chr12_-_19007834 | 0.79 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr22_+_28320792 | 0.78 |
ENSDART00000160562
ENSDART00000164138 |
impg2b
impg2b
|
interphotoreceptor matrix proteoglycan 2b interphotoreceptor matrix proteoglycan 2b |
| chr22_+_35068046 | 0.78 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr10_+_42690374 | 0.75 |
ENSDART00000123496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr11_-_37509001 | 0.74 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr13_-_36911118 | 0.74 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr11_-_37997419 | 0.73 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr20_-_38617766 | 0.72 |
ENSDART00000050474
|
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr20_+_34933183 | 0.72 |
ENSDART00000062738
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr22_-_7129631 | 0.68 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr20_-_46362606 | 0.67 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr23_+_17387325 | 0.67 |
ENSDART00000083947
|
ptk6b
|
PTK6 protein tyrosine kinase 6b |
| chr15_-_21877726 | 0.66 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr23_+_33963619 | 0.65 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr19_-_38830582 | 0.64 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr18_-_48550426 | 0.64 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr20_+_4060839 | 0.64 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr13_-_44630111 | 0.63 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr2_+_21229909 | 0.61 |
ENSDART00000046127
|
vipr1a
|
vasoactive intestinal peptide receptor 1a |
| chr22_-_20386982 | 0.59 |
ENSDART00000089015
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr16_-_12953739 | 0.59 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr12_-_6880694 | 0.59 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr16_-_19890303 | 0.58 |
ENSDART00000147161
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr15_-_23376541 | 0.57 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr12_+_20352400 | 0.57 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr9_+_24008879 | 0.56 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr22_-_10502780 | 0.55 |
ENSDART00000136961
|
ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr24_+_38306010 | 0.54 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr5_+_45139196 | 0.54 |
ENSDART00000113738
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_45323400 | 0.52 |
ENSDART00000148906
ENSDART00000132366 |
emp1
|
epithelial membrane protein 1 |
| chr18_-_1185772 | 0.52 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr21_-_10488379 | 0.52 |
ENSDART00000163878
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr4_+_12615836 | 0.51 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr20_+_28861629 | 0.51 |
ENSDART00000187274
ENSDART00000047826 |
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr15_+_31806303 | 0.50 |
ENSDART00000155902
|
frya
|
furry homolog a (Drosophila) |
| chr4_-_9891874 | 0.48 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr1_-_54765262 | 0.48 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr22_+_5176693 | 0.48 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr18_+_1837668 | 0.48 |
ENSDART00000164210
|
CABZ01079192.1
|
|
| chr23_-_306796 | 0.47 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr22_-_13649588 | 0.47 |
ENSDART00000131877
|
si:ch211-279g13.1
|
si:ch211-279g13.1 |
| chr10_+_11282883 | 0.47 |
ENSDART00000135355
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr11_+_30057762 | 0.46 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr4_-_15420452 | 0.46 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr12_+_22580579 | 0.46 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr6_-_54826061 | 0.45 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr12_+_33395748 | 0.44 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr15_-_22074315 | 0.44 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr20_-_27711970 | 0.44 |
ENSDART00000139637
|
zbtb25
|
zinc finger and BTB domain containing 25 |
| chr5_+_20147830 | 0.43 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr22_+_18477934 | 0.43 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr11_+_38280454 | 0.42 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr24_-_39518599 | 0.42 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr15_-_38154616 | 0.42 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr17_-_19963718 | 0.42 |
ENSDART00000154251
|
chrm3a
|
cholinergic receptor, muscarinic 3a |
| chr2_-_17235891 | 0.41 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr11_-_8126223 | 0.40 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr10_-_35236949 | 0.40 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr10_+_41765944 | 0.40 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr4_+_22343093 | 0.40 |
ENSDART00000023588
|
guca1a
|
guanylate cyclase activator 1A |
| chr16_-_13623928 | 0.39 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr6_+_23887314 | 0.39 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr19_-_44955710 | 0.39 |
ENSDART00000165246
|
CSMD3 (1 of many)
|
si:ch211-233f16.1 |
| chr6_-_6487876 | 0.38 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr5_+_19314574 | 0.37 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr6_+_52804267 | 0.36 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr18_-_43884044 | 0.36 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr16_-_17197546 | 0.36 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr25_+_11456696 | 0.36 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr13_-_40141630 | 0.34 |
ENSDART00000181618
ENSDART00000171583 ENSDART00000192780 |
crtac1b
|
cartilage acidic protein 1b |
| chr15_+_22014029 | 0.33 |
ENSDART00000079504
|
ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr1_-_49947482 | 0.33 |
ENSDART00000143398
|
sgms2
|
sphingomyelin synthase 2 |
| chr17_-_44247707 | 0.32 |
ENSDART00000126097
|
otx2b
|
orthodenticle homeobox 2b |
| chr6_+_39232245 | 0.31 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr11_+_12052791 | 0.30 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr15_+_31344472 | 0.30 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr25_-_27621268 | 0.30 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr25_+_10410620 | 0.30 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr10_-_13343831 | 0.30 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr1_-_44704261 | 0.29 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr20_+_19238382 | 0.29 |
ENSDART00000136757
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr7_-_48263516 | 0.29 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr13_-_25720876 | 0.29 |
ENSDART00000142404
ENSDART00000146487 |
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr13_-_31435137 | 0.28 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr16_+_54263921 | 0.28 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
| chr20_+_28861435 | 0.28 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr5_+_6954162 | 0.28 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr3_-_34296361 | 0.27 |
ENSDART00000181485
ENSDART00000113726 |
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr1_+_52392511 | 0.27 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr21_-_15046065 | 0.25 |
ENSDART00000178507
|
mmp17a
|
matrix metallopeptidase 17a |
| chr8_+_26253252 | 0.25 |
ENSDART00000142031
|
slc26a6
|
solute carrier family 26, member 6 |
| chrM_+_9052 | 0.25 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr6_+_3280939 | 0.24 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chrM_+_9735 | 0.24 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr10_-_10969596 | 0.24 |
ENSDART00000092011
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr10_+_21867307 | 0.24 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr1_-_49947290 | 0.23 |
ENSDART00000141476
|
sgms2
|
sphingomyelin synthase 2 |
| chr22_-_23000815 | 0.23 |
ENSDART00000137111
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr15_-_2184638 | 0.23 |
ENSDART00000135460
|
shox2
|
short stature homeobox 2 |
| chr10_-_10969444 | 0.23 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr10_+_16584382 | 0.22 |
ENSDART00000112039
|
CR790388.1
|
|
| chr19_-_19556778 | 0.22 |
ENSDART00000164060
|
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr10_-_33297864 | 0.20 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr13_+_2625150 | 0.19 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr18_+_19006063 | 0.18 |
ENSDART00000135729
|
slc24a1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr1_+_56755536 | 0.18 |
ENSDART00000157727
|
CABZ01049361.1
|
|
| chr1_-_6028876 | 0.17 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr15_+_15856178 | 0.17 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr7_+_2236317 | 0.16 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr5_-_28109416 | 0.16 |
ENSDART00000042002
|
si:ch211-48m9.1
|
si:ch211-48m9.1 |
| chr9_-_51436377 | 0.15 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr21_+_40417152 | 0.14 |
ENSDART00000171244
|
ssh2b
|
slingshot protein phosphatase 2b |
| chr9_+_17983463 | 0.14 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr25_-_18913336 | 0.13 |
ENSDART00000171010
|
parietopsin
|
parietopsin |
| chr23_-_17484555 | 0.13 |
ENSDART00000187181
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr22_-_20166660 | 0.12 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr13_+_38817871 | 0.10 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr2_+_38147761 | 0.09 |
ENSDART00000135307
|
sall2
|
spalt-like transcription factor 2 |
| chr19_-_12212692 | 0.09 |
ENSDART00000142077
ENSDART00000151599 ENSDART00000140834 ENSDART00000078781 |
znf706
|
zinc finger protein 706 |
| chr8_-_19467011 | 0.09 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr4_+_16885854 | 0.08 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr12_+_34953038 | 0.08 |
ENSDART00000187022
ENSDART00000123988 ENSDART00000027034 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx2a+dlx2b_dlx4a+dlx4b+dlx6a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.4 | 4.3 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.4 | 1.9 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.3 | 1.4 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 2.9 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 1.5 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 1.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 1.4 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.9 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 0.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 0.5 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.7 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.4 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 | 1.0 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.7 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 2.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 2.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 2.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 1.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 3.6 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 0.4 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 3.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 1.6 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.1 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 0.7 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 4.9 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 2.0 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.1 | 0.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 0.3 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.7 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.6 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.5 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 1.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 1.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.0 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.6 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.9 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.7 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.7 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.6 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 1.9 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.6 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 1.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 2.9 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.3 | 1.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 2.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 0.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.8 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 2.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 1.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 5.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.0 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 1.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 1.9 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 1.4 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.3 | 2.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 0.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 1.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 1.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 0.9 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 2.9 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 0.9 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.7 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 3.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 4.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.4 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 2.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 2.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.6 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 3.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.0 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 2.5 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 0.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |