Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
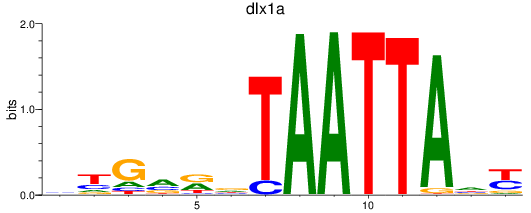
Results for dlx1a
Z-value: 0.87
Transcription factors associated with dlx1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx1a
|
ENSDARG00000013125 | distal-less homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx1a | dr11_v1_chr9_+_3388099_3388099 | -0.43 | 6.3e-02 | Click! |
Activity profile of dlx1a motif
Sorted Z-values of dlx1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_32101202 | 1.97 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr11_-_6877973 | 1.66 |
ENSDART00000160271
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr25_+_14507567 | 1.50 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr7_-_48667056 | 1.43 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr22_-_36530902 | 1.17 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr16_+_54209504 | 1.17 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr23_-_42232124 | 1.16 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr4_+_19700308 | 1.12 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr6_+_21001264 | 1.06 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr20_+_11731039 | 0.96 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr21_-_37973819 | 0.94 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr11_-_33868881 | 0.94 |
ENSDART00000163295
ENSDART00000172633 ENSDART00000171439 |
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr5_+_28497956 | 0.93 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr6_+_7533601 | 0.92 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr6_-_55399214 | 0.92 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr8_-_39822917 | 0.91 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr13_+_7442023 | 0.89 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr21_+_43702016 | 0.89 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr16_+_42471455 | 0.87 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr16_-_27677930 | 0.87 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr4_+_25680480 | 0.86 |
ENSDART00000100737
|
acot17
|
acyl-CoA thioesterase 17 |
| chr13_-_49585385 | 0.86 |
ENSDART00000165868
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr10_-_8046764 | 0.85 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr10_-_35257458 | 0.85 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr1_-_51710225 | 0.84 |
ENSDART00000057601
ENSDART00000152745 |
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr18_+_26422124 | 0.84 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr7_+_29163762 | 0.80 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr17_+_16046132 | 0.79 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr11_-_35171768 | 0.78 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr2_-_57837838 | 0.78 |
ENSDART00000010699
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr5_+_44805269 | 0.77 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr2_-_31833347 | 0.77 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr23_-_16485190 | 0.76 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr23_-_6749820 | 0.76 |
ENSDART00000128772
|
ptges3b
|
prostaglandin E synthase 3b (cytosolic) |
| chr17_-_49412313 | 0.76 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr7_+_69019851 | 0.76 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr20_+_36812368 | 0.76 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr23_-_20126257 | 0.75 |
ENSDART00000005021
|
tktb
|
transketolase b |
| chr17_+_16046314 | 0.75 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr20_-_26822522 | 0.75 |
ENSDART00000146326
ENSDART00000046764 ENSDART00000103234 ENSDART00000143267 |
gmds
|
GDP-mannose 4,6-dehydratase |
| chr15_-_21877726 | 0.75 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr23_+_29885019 | 0.75 |
ENSDART00000167059
|
aurkaip1
|
aurora kinase A interacting protein 1 |
| chr5_-_3574199 | 0.74 |
ENSDART00000060162
|
hspb1
|
heat shock protein, alpha-crystallin-related, 1 |
| chr8_-_31107537 | 0.74 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr7_+_15308219 | 0.73 |
ENSDART00000165683
|
mespba
|
mesoderm posterior ba |
| chr1_-_999556 | 0.73 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr24_+_42004640 | 0.73 |
ENSDART00000171380
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr16_+_54875530 | 0.73 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr8_+_50953776 | 0.72 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr11_+_17984354 | 0.72 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr15_-_43625549 | 0.71 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr5_-_7513082 | 0.71 |
ENSDART00000158913
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
| chr5_-_61624693 | 0.71 |
ENSDART00000141323
|
si:dkey-261j4.4
|
si:dkey-261j4.4 |
| chr12_+_20352400 | 0.70 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr20_-_27225876 | 0.69 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr7_-_17297156 | 0.69 |
ENSDART00000161336
|
nitr11a
|
novel immune-type receptor 11a |
| chr5_-_54672763 | 0.69 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr11_-_40728380 | 0.69 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr25_-_21031007 | 0.68 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr25_+_20715950 | 0.68 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr3_-_34136778 | 0.68 |
ENSDART00000131951
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr5_+_42957503 | 0.67 |
ENSDART00000192885
|
mob1ba
|
MOB kinase activator 1Ba |
| chr18_-_3552414 | 0.67 |
ENSDART00000163762
ENSDART00000165434 ENSDART00000161197 ENSDART00000166841 ENSDART00000170260 |
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr6_+_12922002 | 0.67 |
ENSDART00000080350
ENSDART00000149018 |
cxcr4a
|
chemokine (C-X-C motif) receptor 4a |
| chr10_+_15024772 | 0.67 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr17_+_24618640 | 0.67 |
ENSDART00000092925
|
commd9
|
COMM domain containing 9 |
| chr13_-_12602920 | 0.66 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr7_+_2969661 | 0.66 |
ENSDART00000158382
|
CABZ01007829.1
|
|
| chr14_-_25935167 | 0.66 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr16_+_6021908 | 0.66 |
ENSDART00000163786
|
BX511115.1
|
|
| chr9_+_30421489 | 0.66 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr10_-_21362320 | 0.66 |
ENSDART00000189789
|
avd
|
avidin |
| chr24_+_22485710 | 0.65 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr22_-_10541712 | 0.65 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr10_-_8053385 | 0.64 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr24_+_19415124 | 0.64 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr1_+_54952540 | 0.64 |
ENSDART00000182944
|
r3hcc1l
|
R3H domain and coiled-coil containing 1-like |
| chr1_-_19764038 | 0.64 |
ENSDART00000005733
|
tma16
|
translation machinery associated 16 homolog |
| chr14_-_3381303 | 0.64 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr22_+_16308450 | 0.63 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr1_+_227241 | 0.63 |
ENSDART00000003317
|
tfdp1b
|
transcription factor Dp-1, b |
| chr10_-_21362071 | 0.63 |
ENSDART00000125167
|
avd
|
avidin |
| chr11_+_14321113 | 0.63 |
ENSDART00000039822
ENSDART00000137347 ENSDART00000132997 |
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr25_-_19585010 | 0.63 |
ENSDART00000021340
|
sycp3
|
synaptonemal complex protein 3 |
| chr21_-_32060993 | 0.63 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr6_+_39370587 | 0.63 |
ENSDART00000157165
ENSDART00000155079 |
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr3_+_60716904 | 0.63 |
ENSDART00000168280
|
foxj1a
|
forkhead box J1a |
| chr24_+_14595680 | 0.62 |
ENSDART00000137337
ENSDART00000091784 ENSDART00000136026 |
thtpa
|
thiamine triphosphatase |
| chr23_+_4741543 | 0.62 |
ENSDART00000144761
|
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a |
| chr21_-_26490186 | 0.62 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr3_+_3681116 | 0.62 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr2_+_38373272 | 0.61 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr11_-_3987885 | 0.61 |
ENSDART00000058735
|
glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr22_+_9918872 | 0.60 |
ENSDART00000177953
|
blf
|
bloody fingers |
| chr8_+_23355484 | 0.60 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr2_+_2169337 | 0.60 |
ENSDART00000179939
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr22_-_16154771 | 0.60 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr17_+_30369396 | 0.60 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr19_-_5699703 | 0.60 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr22_+_30447969 | 0.59 |
ENSDART00000193066
|
si:dkey-169i5.4
|
si:dkey-169i5.4 |
| chr5_-_69649044 | 0.59 |
ENSDART00000011192
ENSDART00000170885 |
eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha |
| chr23_+_19813677 | 0.59 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr9_+_35015747 | 0.59 |
ENSDART00000140110
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr18_+_39937225 | 0.58 |
ENSDART00000141136
|
si:ch211-282k23.2
|
si:ch211-282k23.2 |
| chr15_-_15469079 | 0.58 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr20_+_29209615 | 0.58 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr10_+_15025006 | 0.58 |
ENSDART00000145192
ENSDART00000140084 |
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr10_-_8053753 | 0.57 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr3_-_26183699 | 0.57 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr19_-_5669122 | 0.56 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr11_-_25418856 | 0.56 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr2_-_14987282 | 0.56 |
ENSDART00000143057
|
hccsa.2
|
holocytochrome c synthase a, tandem duplicate 2 |
| chr11_+_17984167 | 0.56 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr7_-_13648635 | 0.56 |
ENSDART00000148957
ENSDART00000130967 |
spg21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr9_+_44994214 | 0.55 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr25_-_22363253 | 0.55 |
ENSDART00000073573
ENSDART00000162103 |
ccdc33
|
coiled-coil domain containing 33 |
| chr8_+_23356264 | 0.55 |
ENSDART00000145062
|
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr3_+_30501135 | 0.54 |
ENSDART00000165869
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr7_-_54679595 | 0.54 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr12_-_35830625 | 0.54 |
ENSDART00000180028
|
CU459056.1
|
|
| chr7_+_39418869 | 0.54 |
ENSDART00000169195
|
CT030188.1
|
|
| chr10_+_17714866 | 0.54 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr19_-_25149598 | 0.54 |
ENSDART00000162917
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr17_+_15216022 | 0.53 |
ENSDART00000138831
|
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr10_+_39893439 | 0.53 |
ENSDART00000003435
|
smfn
|
small fragment nuclease |
| chr23_+_34030550 | 0.53 |
ENSDART00000003296
|
sars
|
seryl-tRNA synthetase |
| chr5_+_40485503 | 0.52 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr5_-_41531629 | 0.52 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr1_+_57040472 | 0.52 |
ENSDART00000181365
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr25_-_6292560 | 0.52 |
ENSDART00000153496
|
wdr61
|
WD repeat domain 61 |
| chr2_+_53720028 | 0.51 |
ENSDART00000170799
|
ctnnbl1
|
catenin, beta like 1 |
| chr6_-_33924883 | 0.51 |
ENSDART00000132762
ENSDART00000148142 ENSDART00000142213 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr8_+_31435452 | 0.51 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr17_-_9950911 | 0.51 |
ENSDART00000105117
|
sptssa
|
serine palmitoyltransferase, small subunit A |
| chr17_+_6452511 | 0.51 |
ENSDART00000064694
|
TATDN3
|
Danio rerio TatD DNase domain containing 3-like (LOC571912), mRNA. |
| chr3_+_22442445 | 0.51 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr20_-_22193190 | 0.51 |
ENSDART00000047624
|
tmem165
|
transmembrane protein 165 |
| chr4_+_6032640 | 0.51 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr14_-_36397768 | 0.50 |
ENSDART00000185199
ENSDART00000052562 |
spata4
|
spermatogenesis associated 4 |
| chr10_+_42358426 | 0.50 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr20_+_10538025 | 0.50 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr2_-_9989919 | 0.50 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr10_+_11355841 | 0.50 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr4_-_32180155 | 0.50 |
ENSDART00000164151
|
si:dkey-72l17.6
|
si:dkey-72l17.6 |
| chr20_-_33976305 | 0.49 |
ENSDART00000111022
|
sele
|
selectin E |
| chr21_-_25565392 | 0.49 |
ENSDART00000144917
ENSDART00000180102 |
si:dkey-17e16.10
|
si:dkey-17e16.10 |
| chr1_+_50921266 | 0.49 |
ENSDART00000006538
|
otx2a
|
orthodenticle homeobox 2a |
| chr17_+_24687338 | 0.49 |
ENSDART00000135794
|
selenon
|
selenoprotein N |
| chr2_-_23768818 | 0.49 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr21_+_5862476 | 0.49 |
ENSDART00000065848
|
wdr38
|
WD repeat domain 38 |
| chr14_+_16287968 | 0.49 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr1_+_513986 | 0.48 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr10_+_22771176 | 0.48 |
ENSDART00000192046
|
tmem88a
|
transmembrane protein 88 a |
| chr16_+_23799622 | 0.48 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr15_-_2519640 | 0.48 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr7_-_8712148 | 0.48 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr11_-_21528056 | 0.48 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr19_-_425145 | 0.48 |
ENSDART00000164905
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr18_+_14619544 | 0.48 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr9_-_34937025 | 0.48 |
ENSDART00000137888
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr14_-_41388178 | 0.48 |
ENSDART00000124532
ENSDART00000125016 ENSDART00000169247 |
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr15_-_35098345 | 0.48 |
ENSDART00000181824
|
CR387932.1
|
|
| chr2_+_15048410 | 0.48 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr9_+_41080029 | 0.47 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr1_+_9153141 | 0.47 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr24_-_36175365 | 0.47 |
ENSDART00000065338
|
pak1ip1
|
PAK1 interacting protein 1 |
| chr15_-_18115540 | 0.47 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr5_+_32076109 | 0.47 |
ENSDART00000051357
ENSDART00000144510 |
zmat5
|
zinc finger, matrin-type 5 |
| chr8_+_18011522 | 0.47 |
ENSDART00000136756
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr1_+_58628165 | 0.47 |
ENSDART00000158654
|
si:ch73-221f6.4
|
si:ch73-221f6.4 |
| chr4_+_966061 | 0.47 |
ENSDART00000122535
|
rpap3
|
RNA polymerase II associated protein 3 |
| chr19_+_40026959 | 0.47 |
ENSDART00000123647
|
sfpq
|
splicing factor proline/glutamine-rich |
| chr23_+_37086159 | 0.47 |
ENSDART00000074407
|
cptp
|
ceramide-1-phosphate transfer protein |
| chr9_-_1986014 | 0.46 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr2_-_45331115 | 0.46 |
ENSDART00000158003
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr14_-_33945692 | 0.46 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr19_-_5372572 | 0.46 |
ENSDART00000151326
|
krt17
|
keratin 17 |
| chr25_+_7299488 | 0.46 |
ENSDART00000184836
|
hmg20a
|
high mobility group 20A |
| chr21_-_26028205 | 0.46 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr8_+_2487250 | 0.46 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr19_-_42557416 | 0.45 |
ENSDART00000163217
ENSDART00000128278 ENSDART00000162304 ENSDART00000166556 |
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr11_-_21218172 | 0.45 |
ENSDART00000027532
|
mapkapk2a
|
mitogen-activated protein kinase-activated protein kinase 2a |
| chr7_+_29167744 | 0.45 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr3_-_35541378 | 0.45 |
ENSDART00000183075
ENSDART00000022147 |
ndufab1b
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b |
| chr22_+_16308806 | 0.45 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr1_-_8566567 | 0.45 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr1_+_50987535 | 0.45 |
ENSDART00000140657
|
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr5_+_61459422 | 0.45 |
ENSDART00000050902
|
polr2j
|
polymerase (RNA) II (DNA directed) polypeptide J |
| chr16_+_51154918 | 0.44 |
ENSDART00000058290
|
dhdds
|
dehydrodolichyl diphosphate synthase |
| chr8_+_23826985 | 0.44 |
ENSDART00000187430
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr1_+_52792439 | 0.44 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr3_+_32532645 | 0.44 |
ENSDART00000055312
ENSDART00000150981 |
nosip
|
nitric oxide synthase interacting protein |
| chr10_+_7593185 | 0.44 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr19_+_43780970 | 0.44 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr17_+_24718272 | 0.44 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr5_-_61638125 | 0.44 |
ENSDART00000134314
|
si:dkey-261j4.3
|
si:dkey-261j4.3 |
| chr23_+_31912882 | 0.44 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr7_-_57637779 | 0.44 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr9_+_48123224 | 0.44 |
ENSDART00000141610
|
klhl23
|
kelch-like family member 23 |
| chr22_-_209741 | 0.44 |
ENSDART00000171954
|
ora1
|
olfactory receptor class A related 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.3 | 1.4 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 1.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 1.4 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 0.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 0.7 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.2 | 0.5 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.2 | 1.5 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.2 | 0.5 | GO:1901295 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.2 | 0.9 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 0.8 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.2 | 0.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.2 | 0.9 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.7 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 0.4 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.1 | 0.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 1.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.7 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.6 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.4 | GO:0071047 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 | 0.5 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.5 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.6 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 0.5 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.9 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.6 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 0.5 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.9 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.6 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 0.3 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.5 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.6 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.5 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.6 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.4 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.3 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.1 | 0.2 | GO:0065005 | plasma lipoprotein particle assembly(GO:0034377) protein-lipid complex assembly(GO:0065005) |
| 0.1 | 0.4 | GO:1900221 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.4 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.4 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.7 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.3 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.3 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.5 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.4 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.6 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.7 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.4 | GO:0060584 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.5 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 1.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 | 0.2 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.3 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.8 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.2 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.4 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 1.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 0.7 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.4 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 1.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.3 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.1 | 0.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.4 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.3 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.4 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.6 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.1 | 0.9 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.3 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 1.1 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.5 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.5 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 0.2 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 1.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 0.8 | GO:0051131 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.2 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 0.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 1.0 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.4 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.3 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 1.1 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.4 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.0 | 1.0 | GO:0007286 | spermatid development(GO:0007286) spermatid differentiation(GO:0048515) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.7 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.4 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.3 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.0 | 0.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.4 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.0 | 0.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.1 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.0 | 0.1 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.6 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.2 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.4 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.3 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.6 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 1.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 1.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.9 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0034138 | toll-like receptor 2 signaling pathway(GO:0034134) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.7 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.8 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0071071 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 0.2 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.0 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 2.6 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.4 | GO:0045841 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.1 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.2 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.5 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.3 | GO:1903321 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 1.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 1.5 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.1 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.3 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.4 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 1.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.3 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.5 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 1.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.8 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.3 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.3 | GO:0036297 | oocyte differentiation(GO:0009994) interstrand cross-link repair(GO:0036297) oocyte development(GO:0048599) |
| 0.0 | 0.2 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 0.9 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 0.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 0.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.4 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 0.4 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.1 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.9 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.4 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.7 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.3 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.6 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 0.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.3 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 2.8 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 1.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 0.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 0.7 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 0.7 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.2 | 1.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 0.6 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 0.7 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.4 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.4 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.4 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.4 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.1 | 0.4 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.5 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.1 | 0.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.8 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 2.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.6 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.1 | 0.8 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.5 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.6 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.3 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.6 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.3 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.4 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.1 | 0.5 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.7 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.3 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.2 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.1 | 0.5 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.4 | GO:0071814 | low-density lipoprotein particle binding(GO:0030169) lipoprotein particle binding(GO:0071813) protein-lipid complex binding(GO:0071814) |
| 0.1 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.2 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.3 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.6 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.2 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 1.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0047760 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.2 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.4 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.5 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.0 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.2 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.8 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 3.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 3.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 1.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.7 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.3 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 2.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME MEIOSIS | Genes involved in Meiosis |