Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
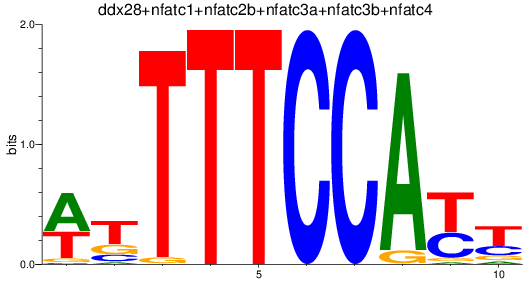
Results for ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
Z-value: 0.89
Transcription factors associated with ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfatc1
|
ENSDARG00000036168 | nuclear factor of activated T cells 1 |
|
nfatc3b
|
ENSDARG00000051729 | nuclear factor of activated T cells 3b |
|
nfatc4
|
ENSDARG00000054162 | nuclear factor of activated T cells 4 |
|
nfatc3a
|
ENSDARG00000076297 | nuclear factor of activated T cells 3a |
|
nfatc2b
|
ENSDARG00000079972 | nuclear factor of activated T cells 2b |
|
ddx28
|
ENSDARG00000112133 | nuclear factor of activated T cells 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfatc2b | dr11_v1_chr6_-_55254786_55254786 | 0.83 | 1.2e-05 | Click! |
| nfatc3b | dr11_v1_chr25_-_36248053_36248053 | 0.81 | 2.7e-05 | Click! |
| nfatc1 | dr11_v1_chr19_+_22216778_22216778 | 0.78 | 9.2e-05 | Click! |
| nfatc4 | dr11_v1_chr2_-_37797577_37797577 | -0.49 | 3.2e-02 | Click! |
| nfatc3a | dr11_v1_chr7_-_34927961_34927961 | 0.34 | 1.5e-01 | Click! |
Activity profile of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4 motif
Sorted Z-values of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_42063906 | 2.87 |
ENSDART00000048893
|
pcbp3
|
poly(rC) binding protein 3 |
| chr19_-_5125663 | 1.97 |
ENSDART00000005547
ENSDART00000132642 |
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr17_+_51499789 | 1.95 |
ENSDART00000187701
|
CABZ01067581.1
|
|
| chr17_+_27456804 | 1.77 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr8_+_22931427 | 1.75 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr20_+_46040666 | 1.75 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr13_+_32740509 | 1.74 |
ENSDART00000076423
ENSDART00000160138 |
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr17_+_33340675 | 1.73 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr12_-_32421046 | 1.73 |
ENSDART00000075567
|
enpp7.1
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 1 |
| chr12_+_46960579 | 1.64 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr16_+_45739193 | 1.60 |
ENSDART00000184852
ENSDART00000156851 ENSDART00000154704 |
paqr6
|
progestin and adipoQ receptor family member VI |
| chr11_-_24191928 | 1.59 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr16_+_45746549 | 1.58 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr3_-_61181018 | 1.50 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr13_+_27073901 | 1.49 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr3_+_9332984 | 1.45 |
ENSDART00000185184
ENSDART00000183535 |
LO018550.2
|
|
| chr21_-_26250443 | 1.39 |
ENSDART00000123713
|
gria1b
|
glutamate receptor, ionotropic, AMPA 1b |
| chr20_-_53624645 | 1.36 |
ENSDART00000083427
ENSDART00000152920 |
slc25a29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr6_-_10233538 | 1.35 |
ENSDART00000182004
ENSDART00000149237 ENSDART00000148876 |
xirp2a
|
xin actin binding repeat containing 2a |
| chr11_-_37359416 | 1.31 |
ENSDART00000159184
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr21_+_11401247 | 1.30 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr24_+_35975398 | 1.30 |
ENSDART00000173058
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr12_-_31103187 | 1.30 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr16_+_4055331 | 1.26 |
ENSDART00000128978
|
CR391998.1
|
|
| chr6_-_48094342 | 1.23 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr3_-_41791178 | 1.22 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr2_-_37353098 | 1.16 |
ENSDART00000056522
|
skila
|
SKI-like proto-oncogene a |
| chr3_-_5067585 | 1.13 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr17_+_30448452 | 1.11 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr9_+_31752628 | 1.10 |
ENSDART00000060054
|
itgbl1
|
integrin, beta-like 1 |
| chr16_-_13613475 | 1.09 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr4_+_61995745 | 1.09 |
ENSDART00000171539
|
CT990567.1
|
|
| chr6_+_10450000 | 1.07 |
ENSDART00000151288
ENSDART00000187431 ENSDART00000192474 ENSDART00000188214 ENSDART00000184766 ENSDART00000190082 |
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr9_+_13682133 | 1.07 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr8_+_37489495 | 1.06 |
ENSDART00000141516
|
fmodb
|
fibromodulin b |
| chr9_-_42989297 | 1.06 |
ENSDART00000126871
|
ttn.2
|
titin, tandem duplicate 2 |
| chr2_+_33541928 | 1.06 |
ENSDART00000162852
|
BX548164.1
|
|
| chr3_+_14463941 | 1.05 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr8_-_21268303 | 1.05 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr12_-_15205087 | 1.04 |
ENSDART00000010068
|
sult1st6
|
sulfotransferase family 1, cytosolic sulfotransferase 6 |
| chr12_-_26064480 | 1.04 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr20_+_6142433 | 1.03 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr23_-_7826849 | 1.01 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr9_-_30190513 | 1.01 |
ENSDART00000137083
|
impg2a
|
interphotoreceptor matrix proteoglycan 2a |
| chr6_+_6491013 | 1.00 |
ENSDART00000140827
|
bcl11ab
|
B cell CLL/lymphoma 11Ab |
| chr8_+_53464216 | 1.00 |
ENSDART00000169514
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr13_-_298454 | 0.99 |
ENSDART00000135782
|
chs1
|
chitin synthase 1 |
| chr4_+_76926059 | 0.97 |
ENSDART00000136192
|
si:dkey-240n22.6
|
si:dkey-240n22.6 |
| chr1_+_19332837 | 0.97 |
ENSDART00000078594
|
tyrp1b
|
tyrosinase-related protein 1b |
| chr22_+_19311411 | 0.97 |
ENSDART00000133234
ENSDART00000138284 |
si:dkey-21e2.16
|
si:dkey-21e2.16 |
| chr15_+_30158652 | 0.97 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr1_-_47089818 | 0.97 |
ENSDART00000132378
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr17_+_13763425 | 0.96 |
ENSDART00000105175
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr12_-_31461219 | 0.95 |
ENSDART00000148954
|
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr13_+_1575276 | 0.94 |
ENSDART00000165987
|
DST
|
dystonin |
| chr12_+_19401854 | 0.94 |
ENSDART00000153415
|
si:dkey-16i5.8
|
si:dkey-16i5.8 |
| chr2_-_13216269 | 0.94 |
ENSDART00000149947
|
bcl2b
|
BCL2, apoptosis regulator b |
| chr6_+_53349966 | 0.92 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr1_-_5455498 | 0.91 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr16_+_14033121 | 0.91 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr2_-_22927958 | 0.90 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr10_-_35236949 | 0.89 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr4_+_19535946 | 0.89 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr15_-_6404443 | 0.88 |
ENSDART00000098038
|
dscamb
|
Down syndrome cell adhesion molecule b |
| chr20_-_39789036 | 0.87 |
ENSDART00000086405
ENSDART00000098253 |
rnf217
|
ring finger protein 217 |
| chr17_+_36627099 | 0.86 |
ENSDART00000154104
|
impg1b
|
interphotoreceptor matrix proteoglycan 1b |
| chr6_-_57539141 | 0.85 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr18_+_3630331 | 0.85 |
ENSDART00000159059
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr4_+_23223881 | 0.85 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr25_+_14087045 | 0.85 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr15_-_34845414 | 0.84 |
ENSDART00000009892
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr2_+_7192966 | 0.84 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr20_+_1996202 | 0.83 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr21_-_21096437 | 0.83 |
ENSDART00000186552
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr5_-_23277939 | 0.83 |
ENSDART00000003514
|
plp1b
|
proteolipid protein 1b |
| chr11_-_37425407 | 0.83 |
ENSDART00000160911
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr1_+_19616799 | 0.80 |
ENSDART00000143077
|
fbxo10
|
F-box protein 10 |
| chr15_+_16525126 | 0.80 |
ENSDART00000193455
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr19_-_4851411 | 0.79 |
ENSDART00000110398
|
fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr17_-_19828505 | 0.79 |
ENSDART00000154745
|
fmn2a
|
formin 2a |
| chr8_-_4327473 | 0.79 |
ENSDART00000134378
|
cux2b
|
cut-like homeobox 2b |
| chr11_-_32723851 | 0.78 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr2_-_22927581 | 0.77 |
ENSDART00000109515
|
myo7bb
|
myosin VIIBb |
| chr10_-_42685512 | 0.76 |
ENSDART00000081347
|
stc1l
|
stanniocalcin 1, like |
| chr12_+_31713239 | 0.76 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr14_-_2369849 | 0.76 |
ENSDART00000180422
ENSDART00000189731 ENSDART00000111748 |
pcdhb
|
protocadherin b |
| chr9_-_23994225 | 0.76 |
ENSDART00000140346
|
col6a3
|
collagen, type VI, alpha 3 |
| chr16_+_46430627 | 0.76 |
ENSDART00000127681
|
rpz6
|
rapunzel 6 |
| chr19_-_15335787 | 0.76 |
ENSDART00000187131
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr12_+_33403694 | 0.75 |
ENSDART00000124083
|
fasn
|
fatty acid synthase |
| chr9_+_54290896 | 0.75 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr9_-_23990416 | 0.75 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr5_+_6755466 | 0.75 |
ENSDART00000163247
ENSDART00000114293 |
areg
|
amphiregulin |
| chr4_-_23083187 | 0.75 |
ENSDART00000009768
|
magi2a
|
membrane associated guanylate kinase, WW and PDZ domain containing 2a |
| chr14_-_448182 | 0.75 |
ENSDART00000180018
|
FAT4
|
FAT atypical cadherin 4 |
| chr9_-_35069645 | 0.74 |
ENSDART00000122679
ENSDART00000077908 ENSDART00000077894 ENSDART00000125536 |
appb
|
amyloid beta (A4) precursor protein b |
| chr14_-_33454595 | 0.74 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr6_-_46398584 | 0.74 |
ENSDART00000193098
|
camk1a
|
calcium/calmodulin-dependent protein kinase Ia |
| chr18_+_41495841 | 0.74 |
ENSDART00000098671
|
si:ch211-203b8.6
|
si:ch211-203b8.6 |
| chr10_+_34623183 | 0.73 |
ENSDART00000114630
|
nbeaa
|
neurobeachin a |
| chr7_+_18176162 | 0.73 |
ENSDART00000109171
|
rce1a
|
Ras converting CAAX endopeptidase 1a |
| chr5_+_51443009 | 0.73 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr19_+_5674907 | 0.73 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr25_+_336503 | 0.73 |
ENSDART00000160395
|
CU929262.1
|
|
| chr2_-_37352514 | 0.72 |
ENSDART00000140498
ENSDART00000186422 |
skila
|
SKI-like proto-oncogene a |
| chr1_+_37391141 | 0.72 |
ENSDART00000083593
ENSDART00000168647 |
sparcl1
|
SPARC-like 1 |
| chr19_+_43392446 | 0.72 |
ENSDART00000147290
|
yrk
|
Yes-related kinase |
| chr20_+_23173710 | 0.71 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr8_+_7144066 | 0.71 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr1_+_38858399 | 0.70 |
ENSDART00000165454
|
CU915762.1
|
|
| chr5_-_30418636 | 0.70 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr24_-_21588914 | 0.70 |
ENSDART00000152054
|
gpr12
|
G protein-coupled receptor 12 |
| chr3_-_30434016 | 0.70 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr21_-_26250744 | 0.69 |
ENSDART00000044735
|
gria1b
|
glutamate receptor, ionotropic, AMPA 1b |
| chr14_-_36345175 | 0.69 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr8_+_36948256 | 0.69 |
ENSDART00000140410
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr20_-_18731268 | 0.69 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr6_-_55254786 | 0.68 |
ENSDART00000113805
|
nfatc2b
|
nuclear factor of activated T cells 2b |
| chr4_+_19534833 | 0.68 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr16_+_48714048 | 0.68 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr2_+_53357953 | 0.67 |
ENSDART00000187577
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr14_-_47314011 | 0.67 |
ENSDART00000178523
|
fstl5
|
follistatin-like 5 |
| chr21_+_40417152 | 0.67 |
ENSDART00000171244
|
ssh2b
|
slingshot protein phosphatase 2b |
| chr2_+_42191592 | 0.67 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr8_-_34051548 | 0.67 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr25_+_27493444 | 0.67 |
ENSDART00000112299
|
gpr37a
|
G protein-coupled receptor 37a |
| chr6_+_10534752 | 0.67 |
ENSDART00000090874
|
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr1_-_38816685 | 0.67 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr2_+_49246099 | 0.66 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr16_+_30117798 | 0.65 |
ENSDART00000135723
ENSDART00000000198 |
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr25_-_17395315 | 0.65 |
ENSDART00000064596
|
cyp2x8
|
cytochrome P450, family 2, subfamily X, polypeptide 8 |
| chr20_-_9273669 | 0.64 |
ENSDART00000175069
|
syt14b
|
synaptotagmin XIVb |
| chr14_+_40874608 | 0.64 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr3_+_22059066 | 0.63 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr2_+_53359234 | 0.62 |
ENSDART00000147581
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr12_-_28537615 | 0.61 |
ENSDART00000067762
|
MYO1D
|
si:ch211-94l19.4 |
| chr9_-_11263228 | 0.61 |
ENSDART00000113847
|
chpfa
|
chondroitin polymerizing factor a |
| chr14_-_2400668 | 0.61 |
ENSDART00000172717
ENSDART00000182882 |
si:ch73-233f7.1
|
si:ch73-233f7.1 |
| chr8_+_50985798 | 0.61 |
ENSDART00000177963
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr7_+_56889879 | 0.61 |
ENSDART00000039810
|
myo9aa
|
myosin IXAa |
| chr11_+_24819624 | 0.61 |
ENSDART00000155514
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr6_-_49078702 | 0.61 |
ENSDART00000135185
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr16_-_12787029 | 0.60 |
ENSDART00000139916
|
foxj2
|
forkhead box J2 |
| chr19_-_46775562 | 0.60 |
ENSDART00000187144
|
abrab
|
actin binding Rho activating protein b |
| chr1_-_59415948 | 0.60 |
ENSDART00000168614
|
si:ch211-188p14.4
|
si:ch211-188p14.4 |
| chr8_-_7637626 | 0.60 |
ENSDART00000123882
ENSDART00000040672 |
mecp2
|
methyl CpG binding protein 2 |
| chr5_-_40910749 | 0.60 |
ENSDART00000083467
ENSDART00000133183 |
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr6_-_40899618 | 0.59 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr6_+_27667359 | 0.59 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr19_-_6193448 | 0.59 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr1_-_21599219 | 0.59 |
ENSDART00000148327
|
adamtsl7
|
ADAMTS-like 7 |
| chr14_+_6429399 | 0.59 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr14_+_4178236 | 0.59 |
ENSDART00000162015
|
slc25a51b
|
solute carrier family 25, member 51b |
| chr7_+_33279108 | 0.58 |
ENSDART00000084530
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr5_-_15851953 | 0.58 |
ENSDART00000173101
|
si:dkey-1k23.3
|
si:dkey-1k23.3 |
| chr10_-_2682198 | 0.58 |
ENSDART00000183727
|
pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr20_-_37522569 | 0.58 |
ENSDART00000177011
ENSDART00000058502 |
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr7_-_11605185 | 0.58 |
ENSDART00000169291
ENSDART00000113904 |
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr3_-_34279109 | 0.57 |
ENSDART00000183255
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr20_-_49681850 | 0.57 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr10_-_23358357 | 0.57 |
ENSDART00000135475
|
cadm2a
|
cell adhesion molecule 2a |
| chr19_+_3056450 | 0.57 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr3_-_28209001 | 0.57 |
ENSDART00000151178
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr13_+_16521898 | 0.57 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr14_-_493029 | 0.56 |
ENSDART00000115093
|
zgc:172215
|
zgc:172215 |
| chr2_-_48966431 | 0.56 |
ENSDART00000147948
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr8_-_38403018 | 0.56 |
ENSDART00000134100
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr4_-_23643272 | 0.56 |
ENSDART00000112301
ENSDART00000133184 |
trhde.2
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 2 |
| chr10_+_35468978 | 0.55 |
ENSDART00000131984
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr20_-_25533739 | 0.55 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr2_-_14390627 | 0.55 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr3_-_32817274 | 0.55 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr16_-_17541890 | 0.55 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr23_-_32892441 | 0.54 |
ENSDART00000147998
|
plxna2
|
plexin A2 |
| chr3_+_12593558 | 0.53 |
ENSDART00000186891
ENSDART00000159252 |
abca3b
|
ATP-binding cassette, sub-family A (ABC1), member 3b |
| chr17_-_45247332 | 0.53 |
ENSDART00000016815
|
ttbk2a
|
tau tubulin kinase 2a |
| chr1_-_49409549 | 0.53 |
ENSDART00000074317
|
GSK3B (1 of many)
|
si:dkeyp-80c12.7 |
| chr25_-_16818978 | 0.53 |
ENSDART00000104140
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr19_-_10243148 | 0.53 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr6_+_36807861 | 0.53 |
ENSDART00000161708
|
si:ch73-29l19.1
|
si:ch73-29l19.1 |
| chr18_+_25546227 | 0.53 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr20_+_31287356 | 0.52 |
ENSDART00000007688
|
slc22a16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr22_+_20461488 | 0.51 |
ENSDART00000135984
|
onecut3a
|
one cut homeobox 3a |
| chr24_+_15897717 | 0.51 |
ENSDART00000105956
|
neto1l
|
neuropilin (NRP) and tolloid (TLL)-like 1, like |
| chr25_+_4837915 | 0.51 |
ENSDART00000168016
|
gnb5a
|
guanine nucleotide binding protein (G protein), beta 5a |
| chr14_+_19258702 | 0.51 |
ENSDART00000187087
ENSDART00000005738 |
slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr10_-_22249444 | 0.51 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr4_-_12102025 | 0.50 |
ENSDART00000048391
ENSDART00000023894 |
braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr13_+_10945337 | 0.50 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr24_+_15670013 | 0.50 |
ENSDART00000185826
|
CU929414.1
|
|
| chr21_-_2814709 | 0.50 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr10_-_29903165 | 0.50 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr17_-_45247151 | 0.50 |
ENSDART00000186230
|
ttbk2a
|
tau tubulin kinase 2a |
| chr14_-_3174570 | 0.49 |
ENSDART00000163585
|
csf1ra
|
colony stimulating factor 1 receptor, a |
| chr4_-_75707991 | 0.49 |
ENSDART00000166358
ENSDART00000160021 |
si:dkey-165o8.2
|
si:dkey-165o8.2 |
| chr23_+_37579107 | 0.49 |
ENSDART00000169376
|
plekhg5b
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5b |
| chr2_-_31634978 | 0.49 |
ENSDART00000135668
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr6_+_45347219 | 0.49 |
ENSDART00000188240
|
LO017951.1
|
|
| chr15_-_20933574 | 0.49 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr14_+_41490821 | 0.49 |
ENSDART00000173319
|
chrnb2l
|
cholinergic receptor, nicotinic, beta 2, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.3 | 1.4 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.3 | 1.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.3 | 1.0 | GO:2000193 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 1.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 1.0 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.2 | 0.7 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 1.0 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.2 | 2.1 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.2 | 1.0 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.2 | 1.6 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.5 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.2 | 1.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 0.6 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.5 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.6 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.5 | GO:0071675 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) regulation of odontogenesis(GO:0042481) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.1 | 0.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.6 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 0.4 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 1.3 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.4 | GO:0021755 | eurydendroid cell differentiation(GO:0021755) |
| 0.1 | 1.0 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.1 | 0.4 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 0.4 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.3 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.4 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.5 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 1.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.3 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 0.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.3 | GO:0044246 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.1 | 0.9 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.9 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.3 | GO:0060405 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.1 | 0.3 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 1.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.3 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.6 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.6 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.3 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.4 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.9 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 1.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.3 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.5 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.6 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 1.1 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.9 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.3 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.9 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.4 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 4.6 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.6 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 2.6 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 1.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 1.2 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.7 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.2 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 2.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.1 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.3 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 1.0 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.7 | GO:0051494 | negative regulation of cytoskeleton organization(GO:0051494) |
| 0.0 | 0.2 | GO:0050930 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.3 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.2 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 4.3 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.7 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.4 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 2.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 2.0 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.0 | 2.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.3 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.2 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0003208 | cardiac ventricle morphogenesis(GO:0003208) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 1.0 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.0 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 1.4 | GO:0006814 | sodium ion transport(GO:0006814) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 1.9 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.0 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.8 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 1.0 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 2.1 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 1.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 2.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 1.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 1.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 3.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 1.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.7 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.4 | 1.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.3 | 1.4 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.3 | 1.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 2.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 0.8 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.2 | 1.7 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.2 | 0.7 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.2 | 1.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 1.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 2.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.2 | 0.6 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.3 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.1 | 1.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.0 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 2.1 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.6 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 1.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.3 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.1 | 0.5 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.6 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 1.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 1.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 2.1 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 1.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.5 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.6 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 3.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 0.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 1.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 0.4 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.4 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME SIGNALLING TO RAS | Genes involved in Signalling to RAS |
| 0.0 | 1.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |