Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
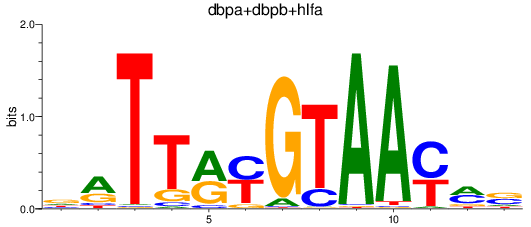
Results for dbpa+dbpb+hlfa
Z-value: 0.97
Transcription factors associated with dbpa+dbpb+hlfa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dbpb
|
ENSDARG00000057652 | D site albumin promoter binding protein b |
|
dbpa
|
ENSDARG00000063014 | D site albumin promoter binding protein a |
|
hlfa
|
ENSDARG00000074752 | HLF transcription factor, PAR bZIP family member a |
|
dbpb
|
ENSDARG00000116703 | D site albumin promoter binding protein b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dbpb | dr11_v1_chr16_-_13613475_13613475 | 0.99 | 1.5e-16 | Click! |
| dbpa | dr11_v1_chr19_-_10196370_10196370 | 0.98 | 9.9e-14 | Click! |
| hlfa | dr11_v1_chr3_-_11624694_11624694 | 0.97 | 2.4e-12 | Click! |
Activity profile of dbpa+dbpb+hlfa motif
Sorted Z-values of dbpa+dbpb+hlfa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_345503 | 5.34 |
ENSDART00000168901
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr5_+_45677781 | 4.18 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr13_-_293250 | 3.84 |
ENSDART00000138581
|
chs1
|
chitin synthase 1 |
| chr21_-_43550120 | 3.78 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr9_-_48736388 | 3.76 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr11_-_29996344 | 3.72 |
ENSDART00000003712
ENSDART00000126110 |
ace2
|
angiotensin I converting enzyme 2 |
| chr2_-_51772438 | 3.41 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr22_+_15315655 | 3.33 |
ENSDART00000141249
|
sult3st3
|
sulfotransferase family 3, cytosolic sulfotransferase 3 |
| chr20_-_25533739 | 3.31 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr20_+_1121458 | 3.22 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr2_+_10134345 | 2.84 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr21_+_43178831 | 2.79 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr17_+_15845765 | 2.53 |
ENSDART00000130881
ENSDART00000074936 |
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr1_-_9231952 | 2.35 |
ENSDART00000166515
|
si:dkeyp-57d7.4
|
si:dkeyp-57d7.4 |
| chr20_-_40755614 | 2.34 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr3_-_28665291 | 2.20 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr12_-_26383242 | 2.17 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr9_+_13682133 | 2.15 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr7_-_41693004 | 2.10 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr24_-_39772045 | 2.05 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr8_+_7975745 | 2.04 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr5_+_51079504 | 2.04 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr21_-_5007109 | 2.03 |
ENSDART00000187042
ENSDART00000097796 ENSDART00000146766 |
rnf165a
|
ring finger protein 165a |
| chr23_-_33361425 | 1.99 |
ENSDART00000031638
|
slc48a1a
|
solute carrier family 48 (heme transporter), member 1a |
| chr20_-_45661049 | 1.99 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr7_+_4162994 | 1.90 |
ENSDART00000172800
|
si:ch211-63p21.1
|
si:ch211-63p21.1 |
| chr25_+_15354095 | 1.89 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr23_-_27345425 | 1.87 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr22_+_15336752 | 1.87 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr7_+_73295890 | 1.87 |
ENSDART00000174331
ENSDART00000174250 |
CABZ01083442.1
|
|
| chr9_-_16109001 | 1.83 |
ENSDART00000053473
|
upp2
|
uridine phosphorylase 2 |
| chr22_+_15323930 | 1.83 |
ENSDART00000142416
|
si:dkey-236e20.3
|
si:dkey-236e20.3 |
| chr14_+_16345003 | 1.83 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr19_+_342094 | 1.83 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr3_+_7763114 | 1.77 |
ENSDART00000057434
|
hook2
|
hook microtubule-tethering protein 2 |
| chr25_-_29415369 | 1.75 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr24_-_38079261 | 1.72 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr9_-_18742704 | 1.70 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr8_+_28900689 | 1.69 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr8_+_1187928 | 1.66 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr25_-_1235457 | 1.64 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr11_-_10770053 | 1.62 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr6_-_39649504 | 1.59 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr14_+_4151379 | 1.59 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr7_-_7823662 | 1.59 |
ENSDART00000167652
|
cxcl8b.3
|
chemokine (C-X-C motif) ligand 8b, duplicate 3 |
| chr23_+_4022620 | 1.57 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr21_-_39058490 | 1.55 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr16_-_33104944 | 1.54 |
ENSDART00000151943
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr18_-_47662696 | 1.51 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr20_+_25563105 | 1.47 |
ENSDART00000063100
|
cyp2p6
|
cytochrome P450, family 2, subfamily P, polypeptide 6 |
| chr16_+_14029283 | 1.46 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr23_+_33963619 | 1.46 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr20_-_1151265 | 1.45 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr8_-_32497815 | 1.44 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr16_+_25245857 | 1.43 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr6_+_8314451 | 1.43 |
ENSDART00000147793
ENSDART00000183688 |
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr5_+_69808763 | 1.38 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr12_+_28574863 | 1.38 |
ENSDART00000153284
|
tbkbp1
|
TBK1 binding protein 1 |
| chr22_+_20720808 | 1.32 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr24_-_24983047 | 1.32 |
ENSDART00000066631
|
slc51a
|
solute carrier family 51, alpha subunit |
| chr7_+_38717624 | 1.31 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr22_+_15331214 | 1.31 |
ENSDART00000136566
|
sult3st4
|
sulfotransferase family 3, cytosolic sulfotransferase 4 |
| chr17_-_6076084 | 1.24 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr22_+_27090136 | 1.21 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr16_+_5408748 | 1.20 |
ENSDART00000160008
ENSDART00000014024 |
plecb
|
plectin b |
| chr6_+_37754763 | 1.19 |
ENSDART00000110770
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr23_-_30960506 | 1.19 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr15_+_26600611 | 1.15 |
ENSDART00000155352
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr5_-_69212184 | 1.15 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr10_-_24371312 | 1.14 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr6_+_28051978 | 1.14 |
ENSDART00000143218
|
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr9_-_1702648 | 1.13 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr16_-_45069882 | 1.13 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr5_-_30074332 | 1.11 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr3_-_36932515 | 1.10 |
ENSDART00000111981
ENSDART00000188470 |
cntnap1
|
contactin associated protein 1 |
| chr21_-_42202792 | 1.10 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chr17_-_6076266 | 1.07 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr12_-_3753131 | 1.07 |
ENSDART00000129668
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr21_-_39931285 | 1.06 |
ENSDART00000180010
ENSDART00000024407 |
tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr11_+_25504215 | 1.06 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr6_-_14292307 | 1.04 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr16_+_46111849 | 1.02 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr13_+_10945337 | 1.02 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr19_+_4990496 | 1.00 |
ENSDART00000151050
ENSDART00000017535 |
zgc:91968
|
zgc:91968 |
| chr19_+_41080240 | 0.99 |
ENSDART00000087295
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr18_-_14941840 | 0.95 |
ENSDART00000091729
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr13_-_10945288 | 0.94 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr20_-_35508805 | 0.94 |
ENSDART00000169538
|
adgrf3b
|
adhesion G protein-coupled receptor F3b |
| chr19_+_9032073 | 0.93 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr11_+_43751263 | 0.92 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr9_-_35069645 | 0.91 |
ENSDART00000122679
ENSDART00000077908 ENSDART00000077894 ENSDART00000125536 |
appb
|
amyloid beta (A4) precursor protein b |
| chr5_-_29488245 | 0.88 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr5_-_16218777 | 0.87 |
ENSDART00000141698
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr19_+_46259619 | 0.86 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr12_+_8003872 | 0.86 |
ENSDART00000180300
ENSDART00000126661 |
rhobtb1
|
Rho-related BTB domain containing 1 |
| chr2_-_54387550 | 0.86 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr21_-_36948 | 0.85 |
ENSDART00000181230
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr19_+_7735157 | 0.83 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr11_+_37251825 | 0.82 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr17_-_7371564 | 0.81 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr19_-_44054930 | 0.80 |
ENSDART00000151084
ENSDART00000150991 ENSDART00000005191 |
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr19_-_22507715 | 0.78 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr5_+_3501859 | 0.77 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr6_-_29612269 | 0.77 |
ENSDART00000104293
|
pex5la
|
peroxisomal biogenesis factor 5-like a |
| chr17_-_34952562 | 0.76 |
ENSDART00000021128
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr6_+_12326267 | 0.76 |
ENSDART00000155101
|
si:dkey-276j7.3
|
si:dkey-276j7.3 |
| chr8_+_8712446 | 0.75 |
ENSDART00000158674
|
elk1
|
ELK1, member of ETS oncogene family |
| chr6_+_8315050 | 0.75 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr16_-_35427060 | 0.74 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr8_-_32497581 | 0.74 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr8_+_12930216 | 0.73 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr1_+_31112436 | 0.72 |
ENSDART00000075340
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr12_+_15002757 | 0.71 |
ENSDART00000135036
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr21_+_7188943 | 0.71 |
ENSDART00000172174
|
agpat2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
| chr11_-_7078392 | 0.71 |
ENSDART00000112156
ENSDART00000188556 |
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr14_-_9199968 | 0.70 |
ENSDART00000146113
|
arhgef9b
|
Cdc42 guanine nucleotide exchange factor (GEF) 9b |
| chr7_+_69528850 | 0.70 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr9_-_53062083 | 0.69 |
ENSDART00000122155
|
zmp:0000000936
|
zmp:0000000936 |
| chr23_-_29812667 | 0.68 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr15_-_22074315 | 0.67 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr6_+_41255485 | 0.67 |
ENSDART00000042683
ENSDART00000186013 |
cadpsb
|
Ca2+-dependent activator protein for secretion b |
| chr9_-_9732212 | 0.65 |
ENSDART00000092884
|
lrrc58b
|
leucine rich repeat containing 58b |
| chr10_-_2527342 | 0.64 |
ENSDART00000184168
|
CU856539.1
|
|
| chr25_+_28893615 | 0.64 |
ENSDART00000156994
ENSDART00000075151 |
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr20_-_15132151 | 0.63 |
ENSDART00000063884
|
si:dkey-239i20.4
|
si:dkey-239i20.4 |
| chr23_+_37323962 | 0.62 |
ENSDART00000102881
|
fam43b
|
family with sequence similarity 43, member B |
| chr13_-_36034582 | 0.62 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr4_-_2727491 | 0.61 |
ENSDART00000141760
ENSDART00000039083 ENSDART00000134442 |
slco1c1
|
solute carrier organic anion transporter family, member 1C1 |
| chr17_-_20979077 | 0.60 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr11_-_45138857 | 0.59 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr20_-_33675676 | 0.57 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr1_-_354115 | 0.57 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr16_-_19568388 | 0.57 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr7_-_46019756 | 0.57 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr5_+_27583117 | 0.56 |
ENSDART00000180340
|
zmat4a
|
zinc finger, matrin-type 4a |
| chr5_-_50638905 | 0.56 |
ENSDART00000180842
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr12_-_46959990 | 0.56 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr16_+_47207691 | 0.56 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr22_-_18491813 | 0.56 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr7_+_30392613 | 0.54 |
ENSDART00000075508
|
lipca
|
lipase, hepatic a |
| chr14_-_12071447 | 0.54 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr10_+_9595575 | 0.52 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr23_+_9522781 | 0.51 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr8_-_29851706 | 0.49 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr3_-_31875138 | 0.49 |
ENSDART00000155298
|
limd2
|
LIM domain containing 2 |
| chr12_-_20373058 | 0.48 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr5_+_32162684 | 0.45 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr15_+_28303161 | 0.44 |
ENSDART00000087926
|
myo1cb
|
myosin Ic, paralog b |
| chr9_-_5318873 | 0.44 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr17_-_31308658 | 0.44 |
ENSDART00000124505
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr7_-_38792543 | 0.43 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr7_+_17716601 | 0.43 |
ENSDART00000173792
ENSDART00000080825 |
rtn3
|
reticulon 3 |
| chr9_-_6927587 | 0.43 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr11_-_40147032 | 0.43 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr17_-_17759138 | 0.40 |
ENSDART00000157128
ENSDART00000123845 |
adck1
|
aarF domain containing kinase 1 |
| chr11_+_23933016 | 0.40 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr7_-_41420226 | 0.39 |
ENSDART00000074155
|
abcf2b
|
ATP-binding cassette, sub-family F (GCN20), member 2b |
| chr22_-_13165186 | 0.39 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr19_+_4990320 | 0.39 |
ENSDART00000147056
|
zgc:91968
|
zgc:91968 |
| chr21_-_20733615 | 0.38 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr17_-_43466317 | 0.38 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr9_-_42418470 | 0.36 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr1_+_2128970 | 0.36 |
ENSDART00000180074
ENSDART00000022019 ENSDART00000098059 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr16_-_7828838 | 0.34 |
ENSDART00000191434
ENSDART00000108653 |
tcaim
|
T cell activation inhibitor, mitochondrial |
| chr12_-_23365737 | 0.33 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr25_-_11088839 | 0.32 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr23_+_45579497 | 0.32 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr8_+_7801060 | 0.31 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr4_-_20181964 | 0.28 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr6_-_41085692 | 0.28 |
ENSDART00000181463
|
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr3_-_336299 | 0.27 |
ENSDART00000105021
|
mhc1zfa
|
major histocompatibility complex class I ZFA |
| chr18_+_507618 | 0.25 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr25_+_7784582 | 0.25 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr25_-_12803723 | 0.24 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr7_+_38278860 | 0.23 |
ENSDART00000016265
|
lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr14_+_21699414 | 0.23 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr5_+_31860043 | 0.22 |
ENSDART00000036235
ENSDART00000140541 |
iscub
|
iron-sulfur cluster assembly enzyme b |
| chr22_+_22004082 | 0.21 |
ENSDART00000148375
|
gna15.2
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 2 |
| chr6_+_50451337 | 0.20 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr15_-_2754056 | 0.20 |
ENSDART00000129380
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr23_-_19486571 | 0.20 |
ENSDART00000009092
|
fam208ab
|
family with sequence similarity 208, member Ab |
| chr3_-_32927516 | 0.20 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr3_+_36972586 | 0.20 |
ENSDART00000102784
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr7_+_38090515 | 0.19 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr9_-_13871935 | 0.18 |
ENSDART00000146597
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr21_-_22715297 | 0.17 |
ENSDART00000065548
|
c1qb
|
complement component 1, q subcomponent, B chain |
| chr9_+_18023288 | 0.17 |
ENSDART00000098355
|
tnfsf11
|
TNF superfamily member 11 |
| chr25_-_16826219 | 0.16 |
ENSDART00000191299
ENSDART00000188504 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr16_-_29397395 | 0.15 |
ENSDART00000130757
|
tlr18
|
toll-like receptor 18 |
| chr23_-_37291793 | 0.15 |
ENSDART00000083281
ENSDART00000187108 |
mul1b
|
mitochondrial E3 ubiquitin protein ligase 1b |
| chr9_+_1654284 | 0.15 |
ENSDART00000062854
|
nfe2l2a
|
nuclear factor, erythroid 2-like 2a |
| chr15_+_15456910 | 0.15 |
ENSDART00000155708
|
zgc:174895
|
zgc:174895 |
| chr22_-_881725 | 0.15 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr3_-_57530323 | 0.14 |
ENSDART00000003066
|
cyth1a
|
cytohesin 1a |
| chr5_-_15494164 | 0.13 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr8_-_43923788 | 0.12 |
ENSDART00000146152
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr22_-_37686966 | 0.10 |
ENSDART00000192217
|
htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr2_+_54387710 | 0.07 |
ENSDART00000127124
|
rab12
|
RAB12, member RAS oncogene family |
| chr3_-_36209936 | 0.06 |
ENSDART00000175208
|
csnk1da
|
casein kinase 1, delta a |
| chr22_-_10121880 | 0.06 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
Network of associatons between targets according to the STRING database.
First level regulatory network of dbpa+dbpb+hlfa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.8 | 3.8 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.5 | 2.0 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.5 | 1.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.4 | 1.4 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 6.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.3 | 2.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 6.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 2.0 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 0.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 0.6 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.2 | 1.8 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.2 | 0.9 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.2 | 1.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 4.2 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.4 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 4.3 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 1.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 1.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 2.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.6 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.1 | 1.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 4.8 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.0 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 1.4 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.1 | 0.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.8 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.8 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 0.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.6 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.1 | 0.7 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.7 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.1 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.7 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.4 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 1.3 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 1.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.8 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 2.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.0 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 1.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.6 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.9 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) regulation of cell junction assembly(GO:1901888) |
| 0.0 | 2.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.1 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 1.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.8 | GO:0001763 | morphogenesis of a branching structure(GO:0001763) |
| 0.0 | 2.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.3 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 1.2 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.5 | 2.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.5 | 3.8 | GO:0030428 | cell septum(GO:0030428) |
| 0.3 | 2.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 1.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 5.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 2.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.9 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 1.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.8 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 3.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.8 | 2.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.5 | 2.2 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.5 | 1.6 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.5 | 3.8 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.5 | 2.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.5 | 3.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.4 | 1.7 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.4 | 5.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.4 | 1.6 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.3 | 1.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.3 | 1.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 2.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 5.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.0 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 2.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.7 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 1.1 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 1.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.4 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 1.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 3.7 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 1.7 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.6 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 1.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.9 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 6.5 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 4.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.7 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.0 | 2.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 1.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 1.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.6 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 1.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 2.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |