Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
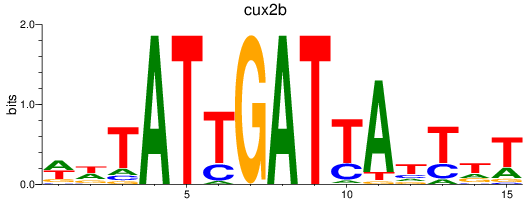
Results for cux2b
Z-value: 0.63
Transcription factors associated with cux2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cux2b
|
ENSDARG00000086345 | cut-like homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cux2b | dr11_v1_chr8_-_4327473_4327473 | -0.14 | 5.6e-01 | Click! |
Activity profile of cux2b motif
Sorted Z-values of cux2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_48039400 | 1.54 |
ENSDART00000166748
ENSDART00000165921 |
csf3b
|
colony stimulating factor 3 (granulocyte) b |
| chr2_-_42958619 | 1.24 |
ENSDART00000144317
|
oc90
|
otoconin 90 |
| chr12_-_20340543 | 1.17 |
ENSDART00000055623
|
hbbe3
|
hemoglobin beta embryonic-3 |
| chr7_+_32695954 | 0.92 |
ENSDART00000184425
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr23_-_45705525 | 0.80 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr21_-_34844059 | 0.69 |
ENSDART00000136402
|
zgc:56585
|
zgc:56585 |
| chr1_+_277731 | 0.67 |
ENSDART00000133431
|
cenpe
|
centromere protein E |
| chr16_+_54210554 | 0.66 |
ENSDART00000172622
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr6_-_17999776 | 0.64 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr19_-_41518922 | 0.63 |
ENSDART00000164483
ENSDART00000062080 |
chrac1
|
chromatin accessibility complex 1 |
| chr3_-_61387273 | 0.63 |
ENSDART00000156479
|
znf1143
|
zinc finger protein 1143 |
| chr16_+_54209504 | 0.63 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr23_+_12454542 | 0.62 |
ENSDART00000182259
ENSDART00000193043 |
si:ch211-153a8.4
|
si:ch211-153a8.4 |
| chr7_-_17297156 | 0.62 |
ENSDART00000161336
|
nitr11a
|
novel immune-type receptor 11a |
| chr17_-_40110782 | 0.59 |
ENSDART00000126929
|
si:dkey-187k19.2
|
si:dkey-187k19.2 |
| chr12_+_16953415 | 0.53 |
ENSDART00000020824
|
pank1b
|
pantothenate kinase 1b |
| chr4_+_5334202 | 0.51 |
ENSDART00000150409
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr20_-_35246150 | 0.50 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr1_+_292545 | 0.50 |
ENSDART00000148261
|
cenpe
|
centromere protein E |
| chr18_-_14677936 | 0.49 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr16_+_29509133 | 0.49 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr21_-_37973819 | 0.49 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr13_-_21650404 | 0.49 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr21_-_2261720 | 0.49 |
ENSDART00000170161
|
si:ch73-299h12.2
|
si:ch73-299h12.2 |
| chr4_+_5334439 | 0.48 |
ENSDART00000180644
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr11_-_35171768 | 0.48 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr9_-_55128839 | 0.48 |
ENSDART00000085135
|
tbl1x
|
transducin beta like 1 X-linked |
| chr3_-_61362398 | 0.47 |
ENSDART00000156177
|
si:dkey-111k8.3
|
si:dkey-111k8.3 |
| chr14_-_970853 | 0.46 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr22_-_3564563 | 0.46 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr18_+_44630415 | 0.46 |
ENSDART00000098540
|
bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr9_-_48371379 | 0.46 |
ENSDART00000115121
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr7_+_1579236 | 0.46 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr15_+_29472065 | 0.45 |
ENSDART00000154343
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr4_+_27018663 | 0.45 |
ENSDART00000180778
|
CR749777.1
|
|
| chr14_+_52481288 | 0.45 |
ENSDART00000169164
ENSDART00000159297 |
tcerg1a
|
transcription elongation regulator 1a (CA150) |
| chr7_-_16597130 | 0.45 |
ENSDART00000144118
|
e2f8
|
E2F transcription factor 8 |
| chr2_+_45300512 | 0.45 |
ENSDART00000144704
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr25_+_3648497 | 0.45 |
ENSDART00000160017
|
CABZ01058261.1
|
|
| chr21_+_45268112 | 0.45 |
ENSDART00000157136
|
tcf7
|
transcription factor 7 |
| chr17_-_2578026 | 0.45 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr22_-_34979139 | 0.44 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr4_+_288633 | 0.44 |
ENSDART00000183304
|
FO834823.1
|
|
| chr4_+_5334707 | 0.43 |
ENSDART00000150620
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr25_+_22320738 | 0.43 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr20_-_16078741 | 0.43 |
ENSDART00000021550
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr21_-_19919918 | 0.42 |
ENSDART00000137307
ENSDART00000142523 ENSDART00000065670 |
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr24_+_38301080 | 0.42 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr1_+_38153944 | 0.42 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr23_-_43486714 | 0.42 |
ENSDART00000169726
|
e2f1
|
E2F transcription factor 1 |
| chr21_+_38855551 | 0.41 |
ENSDART00000171977
|
ddx52
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 |
| chr23_+_4373360 | 0.41 |
ENSDART00000144061
|
ptpdc1b
|
protein tyrosine phosphatase domain containing 1b |
| chr15_+_11814969 | 0.41 |
ENSDART00000127248
|
FO704748.1
|
|
| chr20_+_21391181 | 0.41 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr19_-_10207103 | 0.41 |
ENSDART00000151629
|
znf865
|
zinc finger protein 865 |
| chr7_+_30626378 | 0.41 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr9_+_32073606 | 0.41 |
ENSDART00000184170
ENSDART00000180355 ENSDART00000110204 ENSDART00000123278 |
pikfyve
|
phosphoinositide kinase, FYVE finger containing |
| chr5_+_31779911 | 0.40 |
ENSDART00000098163
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr7_-_5375214 | 0.40 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr12_-_611367 | 0.40 |
ENSDART00000152286
|
wu:fj29h11
|
wu:fj29h11 |
| chr6_+_20647155 | 0.40 |
ENSDART00000193477
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr6_+_18544791 | 0.39 |
ENSDART00000167463
ENSDART00000169599 |
atad5b
|
ATPase family, AAA domain containing 5b |
| chr7_-_16596938 | 0.39 |
ENSDART00000134548
|
e2f8
|
E2F transcription factor 8 |
| chr17_-_2590222 | 0.39 |
ENSDART00000185711
|
CR759892.1
|
|
| chr11_-_22605981 | 0.39 |
ENSDART00000186923
|
myog
|
myogenin |
| chr17_-_15189397 | 0.38 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr16_+_35870456 | 0.38 |
ENSDART00000184321
|
thrap3a
|
thyroid hormone receptor associated protein 3a |
| chr16_-_30564319 | 0.38 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr2_+_21048661 | 0.38 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr19_-_18130567 | 0.38 |
ENSDART00000190659
ENSDART00000022803 |
snx10a
|
sorting nexin 10a |
| chr18_-_15610856 | 0.37 |
ENSDART00000099849
|
arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr9_+_22780901 | 0.37 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
| chr5_-_28767573 | 0.36 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr9_+_13120419 | 0.36 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr16_-_26435431 | 0.36 |
ENSDART00000187526
|
megf8
|
multiple EGF-like-domains 8 |
| chr5_+_37729207 | 0.36 |
ENSDART00000184378
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr3_-_62527675 | 0.36 |
ENSDART00000155048
ENSDART00000064500 |
sox9b
|
SRY (sex determining region Y)-box 9b |
| chr13_+_1726953 | 0.36 |
ENSDART00000103004
|
zmp:0000000760
|
zmp:0000000760 |
| chr21_+_45267589 | 0.36 |
ENSDART00000182963
ENSDART00000155681 ENSDART00000192632 |
tcf7
|
transcription factor 7 |
| chr23_+_21405201 | 0.36 |
ENSDART00000144409
|
iffo2a
|
intermediate filament family orphan 2a |
| chr1_+_16621345 | 0.36 |
ENSDART00000149026
|
pcm1
|
pericentriolar material 1 |
| chr13_+_46941930 | 0.35 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr22_+_18349794 | 0.35 |
ENSDART00000186580
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr14_+_49376011 | 0.34 |
ENSDART00000020961
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr3_+_62339264 | 0.34 |
ENSDART00000174569
|
BX470259.3
|
|
| chr5_-_20205075 | 0.34 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr13_+_45967179 | 0.33 |
ENSDART00000074499
|
olig4
|
oligodendrocyte transcription factor 4 |
| chr8_-_18880399 | 0.33 |
ENSDART00000169043
|
sh3gl1b
|
SH3-domain GRB2-like 1b |
| chr25_+_18964782 | 0.33 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr4_-_77135340 | 0.33 |
ENSDART00000180581
ENSDART00000179901 |
CU467646.7
|
|
| chr25_-_37262220 | 0.32 |
ENSDART00000153789
ENSDART00000155182 |
rfwd3
|
ring finger and WD repeat domain 3 |
| chr19_+_16015881 | 0.32 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr1_+_21563311 | 0.32 |
ENSDART00000147076
ENSDART00000006147 |
primpol
|
primase and polymerase (DNA-directed) |
| chr8_-_53960349 | 0.32 |
ENSDART00000160074
|
cdk11b
|
cyclin-dependent kinase 11B |
| chr10_-_21545091 | 0.32 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr7_+_61480296 | 0.31 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr22_-_7539414 | 0.31 |
ENSDART00000159083
|
BX511034.5
|
|
| chr16_+_28994709 | 0.31 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr19_+_16016038 | 0.31 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr3_+_60721342 | 0.31 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr2_+_35880600 | 0.31 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr10_+_13209580 | 0.31 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr4_+_5832311 | 0.31 |
ENSDART00000121743
ENSDART00000158233 ENSDART00000165187 |
si:ch73-352p4.5
|
si:ch73-352p4.5 |
| chr7_-_16598212 | 0.30 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr23_+_32101202 | 0.30 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr4_-_45115963 | 0.30 |
ENSDART00000137248
|
si:dkey-51d8.6
|
si:dkey-51d8.6 |
| chr2_-_45510699 | 0.30 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr23_+_43424518 | 0.30 |
ENSDART00000022498
|
tti1
|
TELO2 interacting protein 1 |
| chr3_-_61377127 | 0.29 |
ENSDART00000155932
|
si:dkey-111k8.2
|
si:dkey-111k8.2 |
| chr14_+_52413846 | 0.29 |
ENSDART00000160952
|
noa1
|
nitric oxide associated 1 |
| chr2_-_45510223 | 0.29 |
ENSDART00000113058
|
gpsm2
|
G protein signaling modulator 2 |
| chr2_-_34138400 | 0.29 |
ENSDART00000056667
|
cenpl
|
centromere protein L |
| chr21_-_31013817 | 0.29 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr1_+_19930520 | 0.29 |
ENSDART00000158344
|
apbb2b
|
amyloid beta (A4) precursor protein-binding, family B, member 2b |
| chr9_+_12907574 | 0.29 |
ENSDART00000102348
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr19_-_18152942 | 0.29 |
ENSDART00000190182
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr17_-_50045375 | 0.29 |
ENSDART00000184639
|
FO834825.2
|
|
| chr6_-_27891961 | 0.28 |
ENSDART00000155116
|
im:7152348
|
im:7152348 |
| chr17_+_23964132 | 0.28 |
ENSDART00000154823
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr3_-_30488063 | 0.28 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr19_+_627899 | 0.28 |
ENSDART00000148508
|
tert
|
telomerase reverse transcriptase |
| chr19_-_7033636 | 0.28 |
ENSDART00000122815
ENSDART00000123443 ENSDART00000124094 |
daxx
|
death-domain associated protein |
| chr7_-_56831621 | 0.28 |
ENSDART00000182912
|
senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr23_-_33709964 | 0.28 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr8_+_3496204 | 0.28 |
ENSDART00000085993
|
pxnb
|
paxillin b |
| chr13_-_7233811 | 0.27 |
ENSDART00000162026
|
ninl
|
ninein-like |
| chr21_-_11970199 | 0.27 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr7_+_1579510 | 0.27 |
ENSDART00000190525
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr19_+_48102560 | 0.27 |
ENSDART00000164464
|
utp18
|
UTP18 small subunit (SSU) processome component |
| chr7_-_26518086 | 0.27 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr25_+_126174 | 0.27 |
ENSDART00000168913
|
cntn1a
|
contactin 1a |
| chr17_-_43665366 | 0.27 |
ENSDART00000127945
|
egr2a
|
early growth response 2a |
| chr19_+_43256986 | 0.27 |
ENSDART00000182336
|
DGKD
|
diacylglycerol kinase delta |
| chr2_+_20868286 | 0.27 |
ENSDART00000062591
|
odr4
|
odr-4 GPCR localization factor homolog |
| chr8_-_27849770 | 0.27 |
ENSDART00000190196
ENSDART00000181244 |
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr16_-_31790285 | 0.26 |
ENSDART00000184655
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr23_-_43424510 | 0.26 |
ENSDART00000055564
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr9_-_1951144 | 0.26 |
ENSDART00000082355
|
hoxd4a
|
homeobox D4a |
| chr9_+_27329640 | 0.26 |
ENSDART00000111039
|
GTPBP8
|
si:rp71-84d19.3 |
| chr6_-_19926657 | 0.26 |
ENSDART00000157527
ENSDART00000171384 |
ntn1a
|
netrin 1a |
| chr14_-_46259523 | 0.26 |
ENSDART00000172890
|
si:ch211-113d11.8
|
si:ch211-113d11.8 |
| chr19_-_10971230 | 0.26 |
ENSDART00000166196
|
LO018584.1
|
|
| chr17_-_11357851 | 0.26 |
ENSDART00000153915
|
si:ch211-185a18.2
|
si:ch211-185a18.2 |
| chr6_+_3717613 | 0.26 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr2_+_43920461 | 0.26 |
ENSDART00000123673
|
si:ch211-195h23.3
|
si:ch211-195h23.3 |
| chr3_-_58165254 | 0.26 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr11_+_24046179 | 0.26 |
ENSDART00000006703
|
maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr4_-_39171837 | 0.25 |
ENSDART00000179734
|
si:ch211-22k7.9
|
si:ch211-22k7.9 |
| chr15_-_23442891 | 0.25 |
ENSDART00000059376
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr22_+_38978084 | 0.25 |
ENSDART00000025482
|
arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr3_+_18846488 | 0.25 |
ENSDART00000148133
|
tmem104
|
transmembrane protein 104 |
| chr4_-_39111612 | 0.25 |
ENSDART00000150394
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr15_+_32798333 | 0.25 |
ENSDART00000162370
ENSDART00000166525 |
spartb
|
spartin b |
| chr25_+_22017182 | 0.25 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr16_+_53203370 | 0.25 |
ENSDART00000154669
|
si:ch211-269k10.2
|
si:ch211-269k10.2 |
| chr14_-_38843690 | 0.25 |
ENSDART00000183629
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr3_-_16411244 | 0.25 |
ENSDART00000140067
|
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr24_+_37799818 | 0.24 |
ENSDART00000131975
|
telo2
|
TEL2, telomere maintenance 2, homolog (S. cerevisiae) |
| chr23_-_23401305 | 0.24 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr5_+_56268436 | 0.24 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr8_-_10961991 | 0.24 |
ENSDART00000139603
|
trim33
|
tripartite motif containing 33 |
| chr25_+_31122806 | 0.24 |
ENSDART00000067039
|
rassf8a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8a |
| chr4_-_33331681 | 0.24 |
ENSDART00000150319
|
znf1000
|
zinc finger protein 1000 |
| chr2_+_33326522 | 0.24 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr4_+_2655358 | 0.24 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr5_+_53691803 | 0.24 |
ENSDART00000163256
|
taf6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr12_-_4532066 | 0.24 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr5_+_31791001 | 0.24 |
ENSDART00000043010
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr9_+_17423941 | 0.23 |
ENSDART00000112884
ENSDART00000155233 |
kbtbd7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr2_-_24962820 | 0.23 |
ENSDART00000182767
|
hltf
|
helicase-like transcription factor |
| chr12_+_695619 | 0.23 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr4_+_22297839 | 0.23 |
ENSDART00000077707
|
llph
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr21_+_17956856 | 0.23 |
ENSDART00000080431
|
dnai1.2
|
dynein, axonemal, intermediate chain 1, paralog 2 |
| chr15_+_2857556 | 0.23 |
ENSDART00000157758
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr6_-_33916756 | 0.23 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr12_-_30338779 | 0.23 |
ENSDART00000192511
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr4_-_77135076 | 0.23 |
ENSDART00000174184
|
zgc:173770
|
zgc:173770 |
| chr19_-_46018152 | 0.23 |
ENSDART00000159206
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr5_-_2672492 | 0.23 |
ENSDART00000192337
|
CABZ01072548.1
|
|
| chr21_+_20391978 | 0.23 |
ENSDART00000180817
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr21_-_39058490 | 0.23 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr24_-_25461267 | 0.23 |
ENSDART00000105820
|
mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr19_-_47997424 | 0.23 |
ENSDART00000081675
|
ctnnb2
|
catenin, beta 2 |
| chr18_-_12327426 | 0.22 |
ENSDART00000136992
ENSDART00000114024 |
fam107b
|
family with sequence similarity 107, member B |
| chr20_-_13623882 | 0.22 |
ENSDART00000125218
ENSDART00000152499 |
sytl3
|
synaptotagmin-like 3 |
| chr6_+_35052721 | 0.22 |
ENSDART00000191090
ENSDART00000082940 |
uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr2_-_56387041 | 0.22 |
ENSDART00000036240
|
cers4b
|
ceramide synthase 4b |
| chr5_-_39171302 | 0.22 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr7_+_67749251 | 0.22 |
ENSDART00000167562
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr8_-_37101581 | 0.22 |
ENSDART00000185922
|
zgc:162200
|
zgc:162200 |
| chr9_+_20853894 | 0.22 |
ENSDART00000003648
|
wdr3
|
WD repeat domain 3 |
| chr23_+_16807209 | 0.22 |
ENSDART00000141966
|
zgc:114081
|
zgc:114081 |
| chr8_-_21988833 | 0.22 |
ENSDART00000167708
|
nphp4
|
nephronophthisis 4 |
| chr9_-_14143370 | 0.22 |
ENSDART00000108950
|
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr25_+_16356083 | 0.22 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr11_-_34151487 | 0.22 |
ENSDART00000173039
|
atp13a3
|
ATPase 13A3 |
| chr1_+_23784905 | 0.22 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr10_+_16092671 | 0.22 |
ENSDART00000182761
ENSDART00000154835 |
megf10
|
multiple EGF-like-domains 10 |
| chr23_-_478201 | 0.22 |
ENSDART00000140749
|
si:ch73-181d5.4
|
si:ch73-181d5.4 |
| chr23_-_21471022 | 0.22 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cux2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.3 | 1.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.0 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.2 | 0.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.4 | GO:0015893 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.6 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.8 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.3 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.4 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.4 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 1.2 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.3 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 1.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.3 | GO:0009078 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.4 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.1 | 0.2 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.2 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.2 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.1 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.2 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 1.5 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.1 | 0.4 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.2 | GO:0071634 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 | 0.2 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.1 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 1.0 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 0.2 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 0.4 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.1 | 0.3 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.2 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.2 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.1 | 0.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.4 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 1.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.0 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.2 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.3 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.2 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.0 | 1.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.2 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.3 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.4 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0043703 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.7 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.3 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.6 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.3 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.2 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.4 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 0.7 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.2 | 0.6 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 0.6 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.3 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.2 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.2 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.1 | 0.6 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.8 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 1.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 0.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.1 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.4 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.4 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.1 | 0.8 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.3 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.0 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.2 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.1 | 0.2 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 0.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.2 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 1.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.3 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.0 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.0 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |