Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
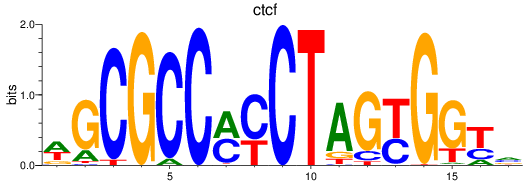
Results for ctcf
Z-value: 2.60
Transcription factors associated with ctcf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ctcf
|
ENSDARG00000056621 | CCCTC-binding factor (zinc finger protein) |
|
ctcf
|
ENSDARG00000113285 | CCCTC-binding factor (zinc finger protein) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ctcf | dr11_v1_chr18_+_22287084_22287188 | -0.36 | 1.3e-01 | Click! |
Activity profile of ctcf motif
Sorted Z-values of ctcf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_22965214 | 9.41 |
ENSDART00000148178
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr3_+_14641962 | 8.67 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr21_-_23331619 | 8.65 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr8_-_22964486 | 7.48 |
ENSDART00000112381
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr18_-_14677936 | 6.97 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr22_+_20135443 | 6.85 |
ENSDART00000143641
|
eef2a.1
|
eukaryotic translation elongation factor 2a, tandem duplicate 1 |
| chr21_-_19918286 | 6.77 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr11_-_41853874 | 6.40 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr13_-_33822550 | 6.13 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr10_-_40826657 | 6.09 |
ENSDART00000076304
|
pcna
|
proliferating cell nuclear antigen |
| chr6_+_60125033 | 5.94 |
ENSDART00000148557
ENSDART00000008224 |
aurka
|
aurora kinase A |
| chr22_+_5135884 | 5.77 |
ENSDART00000141276
|
mydgf
|
myeloid-derived growth factor |
| chr3_+_31093455 | 5.61 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr6_-_42388608 | 5.58 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr7_+_34236238 | 5.30 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr5_+_54497475 | 5.28 |
ENSDART00000158149
ENSDART00000163968 ENSDART00000160446 |
tmem203
|
transmembrane protein 203 |
| chr19_+_20785724 | 5.22 |
ENSDART00000038942
|
adnp2b
|
ADNP homeobox 2b |
| chr5_-_54497319 | 5.03 |
ENSDART00000160492
|
alad
|
aminolevulinate dehydratase |
| chr11_-_25257595 | 4.78 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr2_-_20666920 | 4.75 |
ENSDART00000143437
ENSDART00000114546 ENSDART00000136113 ENSDART00000179247 |
dusp12
|
dual specificity phosphatase 12 |
| chr11_+_2198831 | 4.73 |
ENSDART00000160515
|
hoxc6b
|
homeobox C6b |
| chr2_-_9744081 | 4.69 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr6_+_45934682 | 4.65 |
ENSDART00000103489
|
cenps
|
centromere protein S |
| chr14_-_41468892 | 4.64 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr1_-_8651718 | 4.61 |
ENSDART00000133319
|
actb1
|
actin, beta 1 |
| chr2_-_37896965 | 4.61 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr1_-_22851481 | 4.54 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr17_-_29224908 | 4.50 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr25_+_19733704 | 4.47 |
ENSDART00000145334
ENSDART00000138946 ENSDART00000138851 |
zgc:101783
|
zgc:101783 |
| chr3_-_37148594 | 4.42 |
ENSDART00000140855
|
mlx
|
MLX, MAX dimerization protein |
| chr21_+_43702016 | 4.41 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr12_-_4220713 | 4.40 |
ENSDART00000129427
|
vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr2_+_23701613 | 4.25 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr1_+_218524 | 4.25 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr19_-_33274978 | 4.25 |
ENSDART00000020301
ENSDART00000114714 |
fam92a1
|
family with sequence similarity 92, member A1 |
| chr3_+_37790351 | 4.18 |
ENSDART00000151506
|
si:dkey-260c8.8
|
si:dkey-260c8.8 |
| chr1_+_22851261 | 4.17 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr14_+_48045193 | 4.03 |
ENSDART00000124773
|
ppid
|
peptidylprolyl isomerase D |
| chr19_-_1947403 | 4.01 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr5_-_32396929 | 3.99 |
ENSDART00000023977
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr6_+_47846366 | 3.98 |
ENSDART00000064842
|
padi2
|
peptidyl arginine deiminase, type II |
| chr24_-_13349464 | 3.94 |
ENSDART00000134482
ENSDART00000139212 |
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr1_+_26676758 | 3.90 |
ENSDART00000152299
|
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr4_+_9177997 | 3.88 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr24_-_17400143 | 3.87 |
ENSDART00000134947
|
cul1b
|
cullin 1b |
| chr2_-_25140022 | 3.82 |
ENSDART00000134543
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr21_-_21514176 | 3.74 |
ENSDART00000031205
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr9_+_48819280 | 3.71 |
ENSDART00000112555
|
spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr2_-_37896646 | 3.69 |
ENSDART00000075931
|
hbl1
|
hexose-binding lectin 1 |
| chr3_-_45777226 | 3.68 |
ENSDART00000192849
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr25_+_19734038 | 3.68 |
ENSDART00000067354
|
zgc:101783
|
zgc:101783 |
| chr6_-_35046735 | 3.63 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr1_+_53919110 | 3.62 |
ENSDART00000020680
|
nup133
|
nucleoporin 133 |
| chr13_-_37600600 | 3.60 |
ENSDART00000003888
|
snapc1b
|
small nuclear RNA activating complex, polypeptide 1b |
| chr14_+_6954579 | 3.60 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr16_-_14587332 | 3.59 |
ENSDART00000012479
|
dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr14_-_30557637 | 3.58 |
ENSDART00000183141
|
pfdn6
|
prefoldin subunit 6 |
| chr5_+_41496490 | 3.56 |
ENSDART00000039369
|
fancg
|
Fanconi anemia, complementation group G |
| chr3_+_36671585 | 3.55 |
ENSDART00000159033
|
nde1
|
nudE neurodevelopment protein 1 |
| chr18_+_17021391 | 3.52 |
ENSDART00000100160
|
chtf8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr8_+_20488322 | 3.50 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr12_-_5418340 | 3.48 |
ENSDART00000028043
|
noc3l
|
NOC3-like DNA replication regulator |
| chr6_-_41028698 | 3.45 |
ENSDART00000134293
|
trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr12_-_3077395 | 3.43 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr14_-_30557127 | 3.42 |
ENSDART00000053936
|
pfdn6
|
prefoldin subunit 6 |
| chr14_+_22129096 | 3.39 |
ENSDART00000132514
|
ccng1
|
cyclin G1 |
| chr19_-_19871211 | 3.38 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr22_+_10646928 | 3.36 |
ENSDART00000038465
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr3_+_41647637 | 3.28 |
ENSDART00000050332
|
gna12a
|
guanine nucleotide binding protein (G protein) alpha 12a |
| chr25_-_16755340 | 3.23 |
ENSDART00000124729
ENSDART00000110859 |
ribc2
|
RIB43A domain with coiled-coils 2 |
| chr3_+_24050043 | 3.19 |
ENSDART00000151788
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr12_+_31783066 | 3.17 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr15_-_17406056 | 3.15 |
ENSDART00000146735
|
tubd1
|
tubulin, delta 1 |
| chr22_-_20403194 | 3.15 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr19_-_3886991 | 3.13 |
ENSDART00000162085
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr2_-_26596794 | 3.10 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr19_+_20274944 | 3.05 |
ENSDART00000151237
|
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr23_+_24501918 | 3.04 |
ENSDART00000078824
|
szrd1
|
SUZ RNA binding domain containing 1 |
| chr8_-_11324143 | 3.03 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr24_-_13349802 | 3.02 |
ENSDART00000164729
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr16_+_40340523 | 2.98 |
ENSDART00000102571
|
mettl6
|
methyltransferase like 6 |
| chr15_+_31911989 | 2.97 |
ENSDART00000111472
|
brca2
|
breast cancer 2, early onset |
| chr7_-_60156409 | 2.96 |
ENSDART00000006802
|
cct7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr13_-_35765028 | 2.96 |
ENSDART00000157391
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr1_-_34713710 | 2.92 |
ENSDART00000141569
ENSDART00000102111 ENSDART00000122137 |
tpt1
|
tumor protein, translationally-controlled 1 |
| chr4_+_12931763 | 2.90 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr21_+_25669934 | 2.89 |
ENSDART00000101203
|
sumf2
|
sulfatase modifying factor 2 |
| chr12_-_7639120 | 2.88 |
ENSDART00000126712
ENSDART00000126219 |
ccdc6b
|
coiled-coil domain containing 6b |
| chr4_-_18512550 | 2.87 |
ENSDART00000045639
|
rint1
|
RAD50 interactor 1 |
| chr16_+_28881235 | 2.86 |
ENSDART00000146525
|
chtopb
|
chromatin target of PRMT1b |
| chr7_+_24049776 | 2.81 |
ENSDART00000166559
|
efs
|
embryonal Fyn-associated substrate |
| chr9_+_29985010 | 2.81 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr20_-_30931139 | 2.80 |
ENSDART00000006778
ENSDART00000146376 |
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr2_-_37883256 | 2.79 |
ENSDART00000035685
|
hbl4
|
hexose-binding lectin 4 |
| chr21_+_25120546 | 2.77 |
ENSDART00000149507
|
ddx10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr7_+_66634167 | 2.75 |
ENSDART00000027616
|
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr1_-_45577980 | 2.74 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr2_-_40135942 | 2.73 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr2_+_3823813 | 2.72 |
ENSDART00000103596
ENSDART00000161880 ENSDART00000185408 |
npc1
|
Niemann-Pick disease, type C1 |
| chr14_-_17588345 | 2.72 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr11_+_27364338 | 2.67 |
ENSDART00000186759
|
fbln2
|
fibulin 2 |
| chr22_-_2886937 | 2.67 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr15_+_47525073 | 2.66 |
ENSDART00000067583
|
sidt2
|
SID1 transmembrane family, member 2 |
| chr20_-_36800002 | 2.65 |
ENSDART00000015190
|
ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr6_+_8153813 | 2.64 |
ENSDART00000192436
|
nfil3-3
|
nuclear factor, interleukin 3 regulated, member 3 |
| chr21_+_261490 | 2.57 |
ENSDART00000177919
|
jak2a
|
Janus kinase 2a |
| chr1_+_40613297 | 2.56 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr1_+_22851745 | 2.54 |
ENSDART00000138235
ENSDART00000016488 |
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr2_+_36606019 | 2.54 |
ENSDART00000098415
|
ing5b
|
inhibitor of growth family, member 5b |
| chr25_+_28823952 | 2.53 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr5_-_29514689 | 2.50 |
ENSDART00000126018
ENSDART00000125175 |
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr20_+_49895220 | 2.49 |
ENSDART00000097865
|
ints9
|
integrator complex subunit 9 |
| chr5_-_1869982 | 2.48 |
ENSDART00000055878
|
rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr9_+_21148318 | 2.48 |
ENSDART00000035428
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr16_+_23799622 | 2.48 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr10_-_35186310 | 2.48 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr5_+_64907368 | 2.45 |
ENSDART00000122863
|
ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr18_+_35229115 | 2.42 |
ENSDART00000129624
ENSDART00000184596 |
tbrg1
|
transforming growth factor beta regulator 1 |
| chr23_-_20429741 | 2.40 |
ENSDART00000104334
|
taf13
|
TATA-box binding protein associated factor 13 |
| chr18_+_44649804 | 2.32 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr24_-_17400472 | 2.32 |
ENSDART00000024691
|
cul1b
|
cullin 1b |
| chr18_+_8402129 | 2.28 |
ENSDART00000081154
ENSDART00000171974 |
prpf18
|
PRP18 pre-mRNA processing factor 18 homolog (yeast) |
| chr5_-_58832332 | 2.28 |
ENSDART00000161230
|
arhgef12b
|
Rho guanine nucleotide exchange factor (GEF) 12b |
| chr7_+_52154215 | 2.27 |
ENSDART00000098712
|
TMEM208
|
zgc:77041 |
| chr24_+_37800102 | 2.25 |
ENSDART00000187591
|
telo2
|
TEL2, telomere maintenance 2, homolog (S. cerevisiae) |
| chr6_+_21993613 | 2.24 |
ENSDART00000160750
ENSDART00000083070 |
thumpd3
|
THUMP domain containing 3 |
| chr14_+_31473866 | 2.23 |
ENSDART00000173088
|
ccdc160
|
coiled-coil domain containing 160 |
| chr21_-_32462856 | 2.23 |
ENSDART00000147318
|
zgc:123105
|
zgc:123105 |
| chr8_-_20858676 | 2.18 |
ENSDART00000018515
|
dohh
|
deoxyhypusine hydroxylase/monooxygenase |
| chr24_+_32668675 | 2.16 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr19_-_3906369 | 2.16 |
ENSDART00000160299
ENSDART00000169205 |
si:ch73-281i18.4
|
si:ch73-281i18.4 |
| chr7_-_17816175 | 2.13 |
ENSDART00000091272
ENSDART00000173757 |
ecsit
|
ECSIT signalling integrator |
| chr9_-_12595003 | 2.12 |
ENSDART00000184900
|
tra2b
|
transformer 2 beta homolog |
| chr8_-_17184482 | 2.11 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr5_+_57320113 | 2.10 |
ENSDART00000036331
|
atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr20_+_26002534 | 2.08 |
ENSDART00000147845
|
nrbp1
|
nuclear receptor binding protein 1 |
| chr17_+_21817002 | 2.08 |
ENSDART00000136727
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr8_+_21437908 | 2.08 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr7_-_71384391 | 2.05 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr17_-_735537 | 2.05 |
ENSDART00000166259
|
wars
|
tryptophanyl-tRNA synthetase |
| chr21_+_26539157 | 2.04 |
ENSDART00000021121
|
stx5al
|
syntaxin 5A, like |
| chr10_+_17681074 | 2.03 |
ENSDART00000057500
|
drg1
|
developmentally regulated GTP binding protein 1 |
| chr10_+_11355841 | 1.97 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr12_-_17492852 | 1.93 |
ENSDART00000012421
ENSDART00000138766 ENSDART00000130735 |
minpp1b
|
multiple inositol-polyphosphate phosphatase 1b |
| chr15_+_22435460 | 1.91 |
ENSDART00000031976
|
tmem136a
|
transmembrane protein 136a |
| chr19_-_3916075 | 1.91 |
ENSDART00000161799
|
si:ch73-281i18.7
|
si:ch73-281i18.7 |
| chr9_+_22929675 | 1.87 |
ENSDART00000061299
|
tsn
|
translin |
| chr14_-_30932373 | 1.85 |
ENSDART00000172988
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr25_+_8921425 | 1.84 |
ENSDART00000128591
|
accs
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| chr21_-_21824285 | 1.84 |
ENSDART00000030984
|
wdr73
|
WD repeat domain 73 |
| chr17_-_25303486 | 1.84 |
ENSDART00000162235
|
ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr15_-_16323750 | 1.83 |
ENSDART00000028500
|
nxn
|
nucleoredoxin |
| chr8_+_26007988 | 1.83 |
ENSDART00000193948
ENSDART00000058100 |
xpc
|
xeroderma pigmentosum, complementation group C |
| chr8_-_13364879 | 1.83 |
ENSDART00000063863
|
ccdc124
|
coiled-coil domain containing 124 |
| chr18_-_40753583 | 1.81 |
ENSDART00000026767
|
akt2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr12_-_31726748 | 1.80 |
ENSDART00000153174
|
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr16_+_11585576 | 1.80 |
ENSDART00000172967
|
si:dkey-11o1.6
|
si:dkey-11o1.6 |
| chr7_-_34256374 | 1.78 |
ENSDART00000075176
|
dis3l
|
DIS3 like exosome 3'-5' exoribonuclease |
| chr20_+_9763364 | 1.77 |
ENSDART00000053834
|
psmc6
|
proteasome 26S subunit, ATPase 6 |
| chr11_+_22623284 | 1.77 |
ENSDART00000111850
|
si:ch211-86h15.1
|
si:ch211-86h15.1 |
| chr12_-_33582382 | 1.77 |
ENSDART00000009794
ENSDART00000136617 |
tdrkh
|
tudor and KH domain containing |
| chr6_-_379248 | 1.76 |
ENSDART00000172052
|
si:zfos-169g10.2
|
si:zfos-169g10.2 |
| chr19_-_3925801 | 1.76 |
ENSDART00000129570
ENSDART00000163138 |
si:ch73-281i18.6
|
si:ch73-281i18.6 |
| chr12_-_10567188 | 1.75 |
ENSDART00000144283
|
myof
|
myoferlin |
| chr8_-_51954562 | 1.74 |
ENSDART00000132527
ENSDART00000057315 |
cep78
|
centrosomal protein 78 |
| chr22_-_3344613 | 1.74 |
ENSDART00000165600
|
tbxa2r
|
thromboxane A2 receptor |
| chr2_-_38282079 | 1.72 |
ENSDART00000145808
|
rnf212b
|
si:ch211-10e2.1 |
| chr9_-_12594759 | 1.69 |
ENSDART00000021266
|
tra2b
|
transformer 2 beta homolog |
| chr5_+_32076109 | 1.68 |
ENSDART00000051357
ENSDART00000144510 |
zmat5
|
zinc finger, matrin-type 5 |
| chr21_+_17024002 | 1.65 |
ENSDART00000080628
|
arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr13_+_35528607 | 1.64 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr18_+_3140682 | 1.61 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr21_+_6114709 | 1.61 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr19_+_18903533 | 1.59 |
ENSDART00000157523
ENSDART00000166562 |
slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr5_-_28052883 | 1.57 |
ENSDART00000188791
|
zgc:113436
|
zgc:113436 |
| chr12_-_11294979 | 1.56 |
ENSDART00000148850
|
get4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae) |
| chr17_+_22577472 | 1.56 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr2_+_57149002 | 1.55 |
ENSDART00000168497
|
eef2b
|
eukaryotic translation elongation factor 2b |
| chr14_+_7377552 | 1.55 |
ENSDART00000142158
ENSDART00000141471 |
hars
|
histidyl-tRNA synthetase |
| chr8_+_25900049 | 1.53 |
ENSDART00000124300
ENSDART00000127618 ENSDART00000024009 |
rhoab
|
ras homolog gene family, member Ab |
| chr9_-_14273652 | 1.52 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr19_-_3896514 | 1.51 |
ENSDART00000161346
|
si:ch73-281i18.3
|
si:ch73-281i18.3 |
| chr17_+_25590127 | 1.51 |
ENSDART00000064803
|
iars2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr13_+_35765317 | 1.51 |
ENSDART00000100156
ENSDART00000167650 |
agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr12_+_23991639 | 1.51 |
ENSDART00000003143
|
psme4b
|
proteasome activator subunit 4b |
| chr18_+_17020967 | 1.50 |
ENSDART00000189168
|
chtf8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr13_+_24834199 | 1.50 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr4_+_17245217 | 1.50 |
ENSDART00000184215
ENSDART00000110908 |
casc1
|
cancer susceptibility candidate 1 |
| chr15_-_26887028 | 1.49 |
ENSDART00000156292
|
si:dkey-243i1.1
|
si:dkey-243i1.1 |
| chr19_+_20274264 | 1.48 |
ENSDART00000183513
|
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr11_+_27364059 | 1.47 |
ENSDART00000172883
|
fbln2
|
fibulin 2 |
| chr21_+_17005737 | 1.45 |
ENSDART00000101246
|
vps29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr2_-_42492445 | 1.45 |
ENSDART00000139929
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr5_-_38170996 | 1.44 |
ENSDART00000145805
|
si:ch211-284e13.12
|
si:ch211-284e13.12 |
| chr6_+_11680726 | 1.44 |
ENSDART00000186732
|
calcrlb
|
calcitonin receptor-like b |
| chr24_+_26329018 | 1.44 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr12_-_999762 | 1.43 |
ENSDART00000127003
ENSDART00000084076 ENSDART00000152425 |
mettl9
|
methyltransferase like 9 |
| chr1_+_45958904 | 1.43 |
ENSDART00000108528
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr3_-_30248277 | 1.43 |
ENSDART00000133516
ENSDART00000077029 |
emc10
|
ER membrane protein complex subunit 10 |
| chr9_+_38043337 | 1.42 |
ENSDART00000022574
|
stam2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ctcf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 2.0 | 5.9 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 1.5 | 6.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 1.5 | 4.4 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 1.5 | 4.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 1.4 | 8.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 1.2 | 3.6 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 1.2 | 3.6 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 1.1 | 5.3 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.9 | 3.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.8 | 3.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.8 | 5.6 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.8 | 4.0 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.8 | 3.9 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.8 | 2.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.8 | 3.0 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.7 | 2.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.7 | 2.0 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.7 | 8.6 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.6 | 2.5 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.6 | 2.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.6 | 3.6 | GO:0051299 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.6 | 16.9 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.6 | 2.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.5 | 2.7 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.5 | 1.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.5 | 3.6 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.5 | 9.2 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.5 | 1.0 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.5 | 2.9 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.5 | 1.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.5 | 7.0 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.5 | 1.8 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.4 | 2.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.4 | 2.7 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.4 | 2.3 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.4 | 2.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 | 1.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.3 | 2.1 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.3 | 3.4 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.3 | 5.0 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.3 | 3.4 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.3 | 1.8 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 | 4.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.3 | 2.0 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 3.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.3 | 5.8 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.3 | 6.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 1.8 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.3 | 4.8 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 3.0 | GO:0007530 | sex determination(GO:0007530) |
| 0.2 | 6.2 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.2 | 1.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 5.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 | 5.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 5.1 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.2 | 3.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 2.7 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 1.4 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.2 | 1.0 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 4.9 | GO:0007286 | spermatid development(GO:0007286) spermatid differentiation(GO:0048515) |
| 0.2 | 1.4 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.2 | 0.6 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) positive regulation of DNA binding(GO:0043388) negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.2 | 5.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 0.9 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 2.5 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.2 | 1.0 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 1.6 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.2 | 5.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 3.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 6.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.2 | 1.8 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 1.5 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 1.8 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.6 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 1.7 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.1 | 8.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 2.5 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 3.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 2.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.8 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 2.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 4.9 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.1 | 2.6 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 1.0 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 3.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.4 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.1 | 0.9 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.1 | 1.0 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 1.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 2.9 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.1 | 3.2 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 0.8 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 1.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 3.4 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 2.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 5.1 | GO:0050657 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.1 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 1.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 1.2 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 2.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 1.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 2.5 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 4.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.3 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 2.9 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 3.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.4 | GO:0099560 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) synaptic membrane adhesion(GO:0099560) |
| 0.0 | 1.0 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.9 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 3.0 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 2.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 3.8 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 2.8 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 6.0 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.9 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.8 | GO:0048247 | monocyte chemotaxis(GO:0002548) lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 1.1 | GO:0006473 | protein acetylation(GO:0006473) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 4.1 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.7 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 1.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 5.0 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 0.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 1.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.8 | GO:0098661 | inorganic anion transmembrane transport(GO:0098661) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.3 | 6.7 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 1.2 | 3.6 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 1.1 | 4.4 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 1.1 | 5.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.0 | 5.9 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.9 | 5.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.9 | 4.6 | GO:0097433 | dense body(GO:0097433) |
| 0.8 | 7.0 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.7 | 8.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.6 | 3.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.6 | 2.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.6 | 6.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.6 | 2.2 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.5 | 1.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.5 | 2.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 2.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 6.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 3.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.4 | 7.9 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.3 | 3.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 3.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 1.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 6.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 2.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 1.6 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 1.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.2 | 1.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 0.6 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 1.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.2 | 1.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 2.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 1.8 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.2 | 1.8 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 2.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 2.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 3.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 2.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 2.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 0.8 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 4.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 3.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 2.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 4.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.2 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 1.8 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 4.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.9 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 3.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 10.1 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 4.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 5.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 4.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 6.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.5 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 9.0 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
| 0.0 | 10.9 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.6 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.0 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 3.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.7 | GO:0000502 | proteasome complex(GO:0000502) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 1.2 | 4.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 1.1 | 3.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.9 | 2.8 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.9 | 4.6 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.9 | 3.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.8 | 5.9 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.7 | 3.6 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.6 | 2.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.5 | 8.2 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.5 | 1.9 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.5 | 1.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.5 | 6.8 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.4 | 4.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 7.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.4 | 2.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.4 | 2.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.4 | 1.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.4 | 5.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 3.6 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 2.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.3 | 1.7 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.3 | 0.8 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.3 | 1.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 1.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.3 | 1.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 2.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 0.9 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 1.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 6.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 1.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.2 | 4.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.2 | 2.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 5.1 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.2 | 8.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 7.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 1.4 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.6 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 4.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 2.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 2.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 2.5 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 1.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.0 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 3.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 1.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 8.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 6.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 4.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 1.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 2.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 5.0 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 14.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 3.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 4.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 3.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.8 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 3.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 3.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.8 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 1.2 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 4.4 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.8 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.3 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 4.4 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 3.8 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 4.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 42.5 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 2.1 | GO:0015405 | P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 6.4 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 9.3 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.8 | GO:0015296 | anion:cation symporter activity(GO:0015296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 8.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 13.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 5.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 6.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 4.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.2 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 4.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 4.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 4.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.8 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.5 | 6.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.5 | 4.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 5.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 10.0 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.4 | 2.1 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.3 | 1.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 6.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 1.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 4.4 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.2 | 5.1 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 9.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 2.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 3.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 6.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 1.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 4.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 3.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 7.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 0.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 3.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |