Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
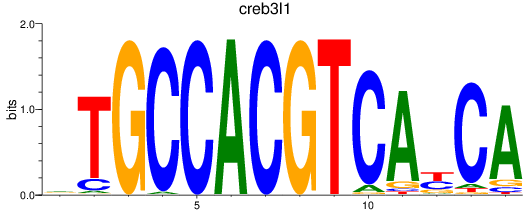
Results for creb3l1
Z-value: 0.42
Transcription factors associated with creb3l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
creb3l1
|
ENSDARG00000015793 | cAMP responsive element binding protein 3-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| creb3l1 | dr11_v1_chr7_+_38897836_38897836 | 0.43 | 6.9e-02 | Click! |
Activity profile of creb3l1 motif
Sorted Z-values of creb3l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_32924669 | 0.75 |
ENSDART00000085219
|
lmo4a
|
LIM domain only 4a |
| chr1_-_7930679 | 0.67 |
ENSDART00000146090
|
si:dkey-79f11.10
|
si:dkey-79f11.10 |
| chr12_-_17479078 | 0.64 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr18_-_34549721 | 0.64 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr3_-_26184018 | 0.59 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr14_-_26411918 | 0.55 |
ENSDART00000020582
|
tmed9
|
transmembrane p24 trafficking protein 9 |
| chr4_+_2637947 | 0.54 |
ENSDART00000130623
|
dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr6_+_13506841 | 0.53 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr4_+_13568469 | 0.52 |
ENSDART00000171235
ENSDART00000136152 |
calua
|
calumenin a |
| chr6_-_9236309 | 0.50 |
ENSDART00000160397
ENSDART00000164603 |
iqcb1
|
IQ motif containing B1 |
| chr15_+_23784842 | 0.49 |
ENSDART00000192889
ENSDART00000138375 |
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr3_-_26183699 | 0.48 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr21_+_10701834 | 0.48 |
ENSDART00000192473
|
lman1
|
lectin, mannose-binding, 1 |
| chr17_-_28797395 | 0.47 |
ENSDART00000134735
|
scfd1
|
sec1 family domain containing 1 |
| chr11_-_2456656 | 0.47 |
ENSDART00000171219
ENSDART00000167938 |
slc2a10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr21_+_10702031 | 0.46 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr22_+_30446987 | 0.44 |
ENSDART00000146471
|
si:dkey-169i5.4
|
si:dkey-169i5.4 |
| chr22_+_31059919 | 0.42 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr15_-_41245962 | 0.42 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr9_+_19529951 | 0.40 |
ENSDART00000125416
|
pknox1.1
|
pbx/knotted 1 homeobox 1.1 |
| chr9_-_42730672 | 0.40 |
ENSDART00000136728
|
fkbp7
|
FK506 binding protein 7 |
| chr18_-_7031409 | 0.39 |
ENSDART00000148485
ENSDART00000005405 |
calub
|
calumenin b |
| chr15_-_18115540 | 0.38 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr3_-_180860 | 0.35 |
ENSDART00000059956
ENSDART00000192506 |
kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr2_-_23172708 | 0.34 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr6_+_3864040 | 0.33 |
ENSDART00000013743
|
gorasp2
|
golgi reassembly stacking protein 2 |
| chr22_-_775776 | 0.31 |
ENSDART00000149749
|
rbbp5
|
retinoblastoma binding protein 5 |
| chr25_-_3034668 | 0.30 |
ENSDART00000053405
|
scamp2
|
secretory carrier membrane protein 2 |
| chr23_+_17878969 | 0.29 |
ENSDART00000154934
ENSDART00000154140 ENSDART00000156331 |
si:ch73-390p7.2
|
si:ch73-390p7.2 |
| chr5_-_48680580 | 0.29 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr18_-_3542435 | 0.29 |
ENSDART00000184017
|
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr20_+_34511678 | 0.28 |
ENSDART00000061588
|
prrx1b
|
paired related homeobox 1b |
| chr16_+_19637384 | 0.26 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr22_-_2886937 | 0.25 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr6_-_50685862 | 0.25 |
ENSDART00000134146
|
mtss1
|
metastasis suppressor 1 |
| chr4_-_17812131 | 0.25 |
ENSDART00000025731
|
spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr1_-_18592068 | 0.24 |
ENSDART00000082063
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr7_-_30177691 | 0.23 |
ENSDART00000046689
|
tmed3
|
transmembrane p24 trafficking protein 3 |
| chr5_-_30516646 | 0.22 |
ENSDART00000014666
|
arcn1a
|
archain 1a |
| chr23_-_25798099 | 0.22 |
ENSDART00000041833
|
fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr21_+_5932140 | 0.21 |
ENSDART00000193767
|
rexo4
|
REX4 homolog, 3'-5' exonuclease |
| chr18_-_18587745 | 0.21 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr4_-_5795309 | 0.21 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr12_+_13348918 | 0.20 |
ENSDART00000181373
|
rnasen
|
ribonuclease type III, nuclear |
| chr8_-_37461835 | 0.20 |
ENSDART00000013446
|
eif2d
|
eukaryotic translation initiation factor 2D |
| chr4_+_11464255 | 0.20 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr3_+_32862730 | 0.20 |
ENSDART00000144939
ENSDART00000125126 ENSDART00000144531 ENSDART00000141717 ENSDART00000137599 |
zgc:162613
|
zgc:162613 |
| chr16_+_24684107 | 0.19 |
ENSDART00000183920
|
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr19_+_20708460 | 0.18 |
ENSDART00000034124
|
rab5aa
|
RAB5A, member RAS oncogene family, a |
| chr7_-_45990386 | 0.18 |
ENSDART00000186008
|
si:ch211-260e23.7
|
si:ch211-260e23.7 |
| chr18_+_7553950 | 0.18 |
ENSDART00000193420
ENSDART00000062143 |
zgc:77650
|
zgc:77650 |
| chr9_-_29497916 | 0.17 |
ENSDART00000060246
|
dnajc3a
|
DnaJ (Hsp40) homolog, subfamily C, member 3a |
| chr6_-_40195510 | 0.16 |
ENSDART00000156156
|
col7a1
|
collagen, type VII, alpha 1 |
| chr10_-_3427589 | 0.15 |
ENSDART00000133452
ENSDART00000037183 |
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr5_-_25066780 | 0.14 |
ENSDART00000002118
ENSDART00000182575 |
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr10_+_31646020 | 0.14 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr7_-_58826164 | 0.14 |
ENSDART00000171095
|
sox32
|
SRY (sex determining region Y)-box 32 |
| chrM_+_9735 | 0.13 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr3_+_26123736 | 0.13 |
ENSDART00000010477
|
hsp70.3
|
heat shock cognate 70-kd protein, tandem duplicate 3 |
| chr15_-_29387446 | 0.12 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr19_+_7549854 | 0.12 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr10_+_15603082 | 0.12 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr20_-_22464250 | 0.11 |
ENSDART00000165904
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr9_+_34230067 | 0.10 |
ENSDART00000139157
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr7_-_45990681 | 0.10 |
ENSDART00000165441
|
si:ch211-260e23.7
|
si:ch211-260e23.7 |
| chr5_+_54685175 | 0.10 |
ENSDART00000115016
|
pmchl
|
pro-melanin-concentrating hormone, like |
| chr20_+_22799641 | 0.09 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr9_+_20554896 | 0.09 |
ENSDART00000144248
|
man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr15_+_32249062 | 0.08 |
ENSDART00000133867
ENSDART00000152545 ENSDART00000082315 ENSDART00000152513 ENSDART00000152139 |
arfip2a
|
ADP-ribosylation factor interacting protein 2a |
| chr4_+_58576146 | 0.08 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr12_-_35582521 | 0.08 |
ENSDART00000162175
ENSDART00000168958 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr4_+_58016732 | 0.08 |
ENSDART00000165777
|
CT027609.1
|
|
| chr20_+_22799857 | 0.07 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr22_-_14346575 | 0.07 |
ENSDART00000136326
|
si:dkeyp-122a9.1
|
si:dkeyp-122a9.1 |
| chr14_-_45595711 | 0.07 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr16_+_13822137 | 0.06 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr20_-_14875308 | 0.06 |
ENSDART00000141290
|
dnm3a
|
dynamin 3a |
| chr20_+_43083745 | 0.06 |
ENSDART00000139014
ENSDART00000153438 |
moxd1l
|
monooxygenase, DBH-like 1, like |
| chr18_-_31423569 | 0.05 |
ENSDART00000185028
|
CU570893.1
|
|
| chr5_-_39474235 | 0.05 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr19_+_44039849 | 0.05 |
ENSDART00000086040
|
lrrc14b
|
leucine rich repeat containing 14B |
| chr1_-_49649122 | 0.04 |
ENSDART00000134399
|
slkb
|
STE20-like kinase b |
| chr9_+_45789887 | 0.04 |
ENSDART00000135202
|
si:dkey-34f9.3
|
si:dkey-34f9.3 |
| chr13_-_9875538 | 0.04 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr18_-_7481036 | 0.03 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr22_-_1168291 | 0.03 |
ENSDART00000167724
|
si:ch1073-181h11.4
|
si:ch1073-181h11.4 |
| chr22_-_10482253 | 0.03 |
ENSDART00000143164
|
aspn
|
asporin (LRR class 1) |
| chr2_+_5948534 | 0.02 |
ENSDART00000124324
ENSDART00000176461 |
slc1a7a
|
solute carrier family 1 (glutamate transporter), member 7a |
| chr17_+_33767890 | 0.02 |
ENSDART00000193177
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr7_+_13491452 | 0.00 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
Network of associatons between targets according to the STRING database.
First level regulatory network of creb3l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 0.6 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 0.6 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.5 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.5 | GO:0036372 | opsin transport(GO:0036372) |
| 0.1 | 0.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.4 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 1.2 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 1.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.5 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |