Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
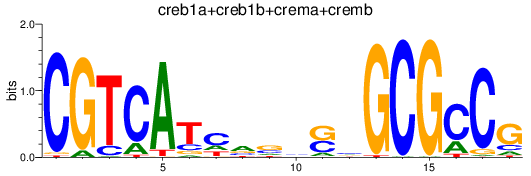
Results for creb1a+creb1b+crema+cremb
Z-value: 8.02
Transcription factors associated with creb1a+creb1b+crema+cremb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
crema
|
ENSDARG00000023217 | cAMP responsive element modulator a |
|
creb1a
|
ENSDARG00000053586 | cAMP responsive element binding protein 1a |
|
creb1b
|
ENSDARG00000093420 | cAMP responsive element binding protein 1b |
|
cremb
|
ENSDARG00000102899 | cAMP responsive element modulator b |
|
cremb
|
ENSDARG00000109505 | cAMP responsive element modulator b |
|
cremb
|
ENSDARG00000116064 | cAMP responsive element modulator b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| crema | dr11_v1_chr2_-_43168292_43168292 | -0.97 | 2.5e-12 | Click! |
| cremb | dr11_v1_chr24_+_31361407_31361407 | 0.88 | 8.3e-07 | Click! |
| creb1a | dr11_v1_chr1_-_33557915_33557915 | 0.77 | 1.0e-04 | Click! |
| creb1b | dr11_v1_chr9_-_28275600_28275600 | 0.36 | 1.4e-01 | Click! |
Activity profile of creb1a+creb1b+crema+cremb motif
Sorted Z-values of creb1a+creb1b+crema+cremb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_42556555 | 20.74 |
ENSDART00000168536
|
znf1053
|
zinc finger protein 1053 |
| chr4_-_56898328 | 18.15 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr4_-_57001458 | 17.68 |
ENSDART00000158660
|
si:ch211-161m3.1
|
si:ch211-161m3.1 |
| chr15_+_23722620 | 17.10 |
ENSDART00000011447
|
sae1
|
SUMO1 activating enzyme subunit 1 |
| chr19_-_3056235 | 16.77 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr4_-_61651223 | 16.71 |
ENSDART00000172688
|
si:dkey-26i24.1
|
si:dkey-26i24.1 |
| chr4_-_50544015 | 16.70 |
ENSDART00000155155
|
zgc:174704
|
zgc:174704 |
| chr10_-_23099809 | 16.66 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr4_-_32180155 | 15.75 |
ENSDART00000164151
|
si:dkey-72l17.6
|
si:dkey-72l17.6 |
| chr4_+_45504471 | 15.70 |
ENSDART00000150399
|
si:dkey-256i11.2
|
si:dkey-256i11.2 |
| chr4_-_56068511 | 15.67 |
ENSDART00000168345
|
znf1133
|
zinc finger protein 1133 |
| chr4_+_33461796 | 15.65 |
ENSDART00000150445
|
si:dkey-247i3.1
|
si:dkey-247i3.1 |
| chr4_+_30454496 | 15.50 |
ENSDART00000164555
|
si:dkey-199m13.4
|
si:dkey-199m13.4 |
| chr4_-_43280244 | 15.39 |
ENSDART00000150762
|
si:dkeyp-53e4.1
|
si:dkeyp-53e4.1 |
| chr4_-_49952636 | 15.22 |
ENSDART00000157941
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr3_-_62194512 | 14.87 |
ENSDART00000074174
|
tbl3
|
transducin (beta)-like 3 |
| chr4_-_33331681 | 14.85 |
ENSDART00000150319
|
znf1000
|
zinc finger protein 1000 |
| chr4_-_61650771 | 14.84 |
ENSDART00000168593
|
si:dkey-26i24.1
|
si:dkey-26i24.1 |
| chr19_-_27339670 | 14.81 |
ENSDART00000139323
|
znrd1
|
zinc ribbon domain containing 1 |
| chr20_+_47143900 | 14.48 |
ENSDART00000153360
|
si:dkeyp-104f11.6
|
si:dkeyp-104f11.6 |
| chr4_+_62184754 | 14.31 |
ENSDART00000168844
|
si:dkeyp-35e5.9
|
si:dkeyp-35e5.9 |
| chr20_-_36679233 | 14.27 |
ENSDART00000062908
|
rpl7l1
|
ribosomal protein L7-like 1 |
| chr4_-_57530817 | 14.25 |
ENSDART00000158435
|
zgc:173702
|
zgc:173702 |
| chr4_+_49073583 | 14.24 |
ENSDART00000170022
|
zgc:173705
|
zgc:173705 |
| chr4_-_69615167 | 14.23 |
ENSDART00000171108
|
si:ch211-120c15.3
|
si:ch211-120c15.3 |
| chr3_+_32651697 | 14.17 |
ENSDART00000055338
|
thoc6
|
THO complex 6 |
| chr22_-_17606575 | 14.11 |
ENSDART00000183951
|
gpx4a
|
glutathione peroxidase 4a |
| chr4_-_55459037 | 14.10 |
ENSDART00000172184
|
znf569l
|
zinc finger protein 569, like |
| chr4_-_45301719 | 13.81 |
ENSDART00000150282
|
si:ch211-162i8.2
|
si:ch211-162i8.2 |
| chr6_+_3716666 | 13.79 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr18_-_5850683 | 13.56 |
ENSDART00000082087
|
nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr5_+_50869091 | 13.39 |
ENSDART00000083294
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr4_+_69823638 | 13.31 |
ENSDART00000165786
|
znf1087
|
zinc finger protein 1087 |
| chr4_+_59320243 | 13.29 |
ENSDART00000150611
|
znf1084
|
zinc finger protein 1084 |
| chr6_-_9236309 | 13.20 |
ENSDART00000160397
ENSDART00000164603 |
iqcb1
|
IQ motif containing B1 |
| chr4_-_42294516 | 13.16 |
ENSDART00000133558
|
si:dkey-4e4.1
|
si:dkey-4e4.1 |
| chr4_-_49720964 | 13.14 |
ENSDART00000156103
|
znf1057
|
zinc finger protein 1057 |
| chr4_+_64577406 | 13.07 |
ENSDART00000159754
|
si:ch211-223a21.4
|
si:ch211-223a21.4 |
| chr4_-_70488123 | 12.94 |
ENSDART00000169266
|
si:dkeyp-44b5.5
|
si:dkeyp-44b5.5 |
| chr2_-_58374842 | 12.92 |
ENSDART00000111651
|
AL954696.1
|
|
| chr4_+_58576146 | 12.89 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr4_+_59711338 | 12.77 |
ENSDART00000150849
|
si:dkey-149m13.4
|
si:dkey-149m13.4 |
| chr4_+_38103421 | 12.77 |
ENSDART00000164164
|
znf1016
|
zinc finger protein 1016 |
| chr4_-_32471434 | 12.71 |
ENSDART00000152013
ENSDART00000183621 |
znf1085
|
zinc finger protein 1085 |
| chr4_-_52621665 | 12.64 |
ENSDART00000137064
|
si:dkeyp-104f11.6
|
si:dkeyp-104f11.6 |
| chr4_-_45301250 | 12.31 |
ENSDART00000181753
|
si:ch211-162i8.2
|
si:ch211-162i8.2 |
| chr3_-_16413606 | 12.26 |
ENSDART00000127309
ENSDART00000017172 ENSDART00000136465 |
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr8_+_13064750 | 12.25 |
ENSDART00000039878
|
sap30bp
|
SAP30 binding protein |
| chr15_+_823437 | 12.13 |
ENSDART00000155608
|
si:dkey-7i4.7
|
si:dkey-7i4.7 |
| chr19_-_8536474 | 12.03 |
ENSDART00000139715
|
dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr12_-_30043809 | 11.89 |
ENSDART00000150220
ENSDART00000075889 ENSDART00000148847 |
trub1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr23_-_43424510 | 11.76 |
ENSDART00000055564
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr4_+_63818718 | 11.76 |
ENSDART00000161177
|
si:dkey-30f3.2
|
si:dkey-30f3.2 |
| chr4_-_64142389 | 11.48 |
ENSDART00000172126
|
BX914205.3
|
|
| chr13_-_36141486 | 11.47 |
ENSDART00000044115
|
med6
|
mediator complex subunit 6 |
| chr4_+_58016732 | 11.42 |
ENSDART00000165777
|
CT027609.1
|
|
| chr4_+_59234719 | 11.38 |
ENSDART00000170724
ENSDART00000192143 ENSDART00000109914 |
znf1086
|
zinc finger protein 1086 |
| chr4_-_30712160 | 11.37 |
ENSDART00000141715
|
si:dkey-16p19.5
|
si:dkey-16p19.5 |
| chr19_-_27339844 | 11.34 |
ENSDART00000052358
ENSDART00000148238 ENSDART00000147661 ENSDART00000137346 |
znrd1
|
zinc ribbon domain containing 1 |
| chr4_+_58057168 | 11.29 |
ENSDART00000161097
|
si:dkey-57k17.1
|
si:dkey-57k17.1 |
| chr1_+_19083501 | 11.23 |
ENSDART00000002886
ENSDART00000131948 |
exosc9
|
exosome component 9 |
| chr4_-_36144500 | 11.20 |
ENSDART00000170896
|
znf992
|
zinc finger protein 992 |
| chr5_-_26199505 | 11.19 |
ENSDART00000132950
|
rad17
|
RAD17 checkpoint clamp loader component |
| chr4_-_52621232 | 11.19 |
ENSDART00000124451
|
si:dkeyp-104f11.6
|
si:dkeyp-104f11.6 |
| chr17_-_49481672 | 11.16 |
ENSDART00000166394
|
fcf1
|
FCF1 rRNA-processing protein |
| chr4_+_45504938 | 11.14 |
ENSDART00000145958
|
si:dkey-256i11.2
|
si:dkey-256i11.2 |
| chr8_-_32354677 | 11.13 |
ENSDART00000138268
ENSDART00000133245 ENSDART00000179677 ENSDART00000174450 |
ipo11
|
importin 11 |
| chr4_+_33373100 | 10.87 |
ENSDART00000150417
|
znf1090
|
zinc finger protein 1090 |
| chr4_+_54798291 | 10.85 |
ENSDART00000165113
ENSDART00000109624 |
si:dkeyp-82b4.6
|
si:dkeyp-82b4.6 |
| chr4_-_59159690 | 10.82 |
ENSDART00000164706
|
znf1149
|
zinc finger protein 1149 |
| chr9_+_145093 | 10.76 |
ENSDART00000162764
|
rrp1
|
ribosomal RNA processing 1 |
| chr4_+_59748607 | 10.76 |
ENSDART00000108499
|
znf1068
|
zinc finger protein 1068 |
| chr4_-_50722304 | 10.73 |
ENSDART00000150480
|
znf1110
|
zinc finger protein 1110 |
| chr4_+_71819777 | 10.70 |
ENSDART00000171070
|
znf1017
|
zinc finger protein 1017 |
| chr4_+_63769987 | 10.63 |
ENSDART00000168878
|
znf1048
|
zinc finger protein 1048 |
| chr4_+_56315530 | 10.62 |
ENSDART00000169288
|
zgc:174275
|
zgc:174275 |
| chr8_-_28274251 | 10.58 |
ENSDART00000050671
|
rap1aa
|
RAP1A, member of RAS oncogene family a |
| chr4_-_64336954 | 10.50 |
ENSDART00000157986
|
znf1147
|
zinc finger protein 1147 |
| chr7_-_40657831 | 10.50 |
ENSDART00000084153
|
nom1
|
nucleolar protein with MIF4G domain 1 |
| chr4_+_63818212 | 10.44 |
ENSDART00000164929
|
si:dkey-30f3.2
|
si:dkey-30f3.2 |
| chr4_-_69917450 | 10.41 |
ENSDART00000168915
|
znf1076
|
zinc finger protein 1076 |
| chr4_-_57994891 | 10.35 |
ENSDART00000168657
|
znf1097
|
zinc finger protein 1097 |
| chr4_+_69510337 | 10.34 |
ENSDART00000165995
|
znf1025
|
zinc finger protein 1025 |
| chr4_-_69127091 | 10.31 |
ENSDART00000136092
|
si:ch211-209j12.3
|
si:ch211-209j12.3 |
| chr4_+_61865790 | 10.30 |
ENSDART00000166793
|
si:dkey-146c18.5
|
si:dkey-146c18.5 |
| chr4_+_43408004 | 10.30 |
ENSDART00000150476
|
si:dkeyp-53e4.2
|
si:dkeyp-53e4.2 |
| chr10_+_28306749 | 10.23 |
ENSDART00000142016
|
ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr4_-_77218637 | 10.19 |
ENSDART00000174325
|
psmb10
|
proteasome subunit beta 10 |
| chr5_+_72194444 | 10.19 |
ENSDART00000165436
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr4_+_64577910 | 10.11 |
ENSDART00000128164
|
si:ch211-223a21.4
|
si:ch211-223a21.4 |
| chr4_-_73488406 | 10.11 |
ENSDART00000115002
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr4_-_35900331 | 10.10 |
ENSDART00000164413
|
zgc:174653
|
zgc:174653 |
| chr4_-_50158550 | 10.00 |
ENSDART00000121664
|
CR356227.1
|
|
| chr16_+_25074029 | 9.97 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr4_+_62262253 | 9.93 |
ENSDART00000166022
|
si:dkeyp-35e5.10
|
si:dkeyp-35e5.10 |
| chr4_-_43279775 | 9.90 |
ENSDART00000183160
|
si:dkeyp-53e4.1
|
si:dkeyp-53e4.1 |
| chr4_+_69863019 | 9.90 |
ENSDART00000168400
|
znf1117
|
zinc finger protein 1117 |
| chr4_-_51045333 | 9.84 |
ENSDART00000150560
|
znf1026
|
zinc finger protein 1026 |
| chr23_-_17450746 | 9.80 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr11_-_12802123 | 9.75 |
ENSDART00000104143
|
txlng
|
taxilin gamma |
| chr4_+_28962774 | 9.73 |
ENSDART00000150629
|
si:dkey-23a23.3
|
si:dkey-23a23.3 |
| chr4_-_25515154 | 9.73 |
ENSDART00000186524
|
rbm17
|
RNA binding motif protein 17 |
| chr4_+_41528590 | 9.71 |
ENSDART00000164004
|
znf1094
|
zinc finger protein 1094 |
| chr4_-_65090543 | 9.67 |
ENSDART00000167888
|
si:ch211-194h1.2
|
si:ch211-194h1.2 |
| chr13_-_31370184 | 9.66 |
ENSDART00000034829
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr4_+_64147241 | 9.61 |
ENSDART00000163509
|
znf1089
|
zinc finger protein 1089 |
| chr9_+_22929675 | 9.56 |
ENSDART00000061299
|
tsn
|
translin |
| chr4_-_44673017 | 9.56 |
ENSDART00000156670
|
si:dkey-7j22.4
|
si:dkey-7j22.4 |
| chr4_-_59057046 | 9.42 |
ENSDART00000150249
|
znf1132
|
zinc finger protein 1132 |
| chr4_-_44872722 | 9.41 |
ENSDART00000150621
|
BX649307.1
|
|
| chr4_+_29909870 | 9.39 |
ENSDART00000150324
|
znf1111
|
zinc finger protein 1111 |
| chr14_-_21932403 | 9.33 |
ENSDART00000054420
|
rad9a
|
RAD9 checkpoint clamp component A |
| chr4_-_50788075 | 9.33 |
ENSDART00000150302
|
znf1045
|
zinc finger protein 1045 |
| chr4_-_43185261 | 9.28 |
ENSDART00000150380
|
si:dkey-16p6.2
|
si:dkey-16p6.2 |
| chr4_+_28418829 | 9.14 |
ENSDART00000111603
|
znf1041
|
zinc finger protein 1041 |
| chr4_-_67143082 | 9.14 |
ENSDART00000162586
|
CT573263.1
|
|
| chr4_+_53183373 | 9.04 |
ENSDART00000176617
|
znf1037
|
zinc finger protein 1037 |
| chr8_+_49065348 | 9.03 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr13_-_31687925 | 8.95 |
ENSDART00000085989
|
trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) |
| chr4_+_69559692 | 8.93 |
ENSDART00000164383
|
znf993
|
zinc finger protein 993 |
| chr23_+_43424518 | 8.92 |
ENSDART00000022498
|
tti1
|
TELO2 interacting protein 1 |
| chr4_-_42074960 | 8.92 |
ENSDART00000164406
|
zgc:174696
|
zgc:174696 |
| chr4_-_45115963 | 8.82 |
ENSDART00000137248
|
si:dkey-51d8.6
|
si:dkey-51d8.6 |
| chr8_-_28274552 | 8.77 |
ENSDART00000131580
|
rap1aa
|
RAP1A, member of RAS oncogene family a |
| chr4_-_63769147 | 8.76 |
ENSDART00000189919
|
znf1098
|
zinc finger protein 1098 |
| chr4_+_29344957 | 8.73 |
ENSDART00000185164
|
CR847895.1
|
|
| chr3_+_9588705 | 8.71 |
ENSDART00000172543
ENSDART00000104875 |
trap1
|
TNF receptor-associated protein 1 |
| chr4_+_60381608 | 8.69 |
ENSDART00000167351
|
znf1130
|
zinc finger protein 1130 |
| chr8_+_12118097 | 8.66 |
ENSDART00000081819
|
endog
|
endonuclease G |
| chr11_-_45433419 | 8.65 |
ENSDART00000163923
ENSDART00000075612 |
rfc4
|
replication factor C (activator 1) 4 |
| chr4_+_64102312 | 8.64 |
ENSDART00000157859
|
znf1103
|
zinc finger protein 1103 |
| chr10_+_1681518 | 8.63 |
ENSDART00000018532
|
triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr15_+_846768 | 8.47 |
ENSDART00000155633
ENSDART00000191235 |
si:dkey-7i4.5
si:dkey-77f5.8
|
si:dkey-7i4.5 si:dkey-77f5.8 |
| chr7_-_7797654 | 8.47 |
ENSDART00000084503
ENSDART00000192779 ENSDART00000173079 |
trmt10b
|
tRNA methyltransferase 10B |
| chr17_+_13007307 | 8.45 |
ENSDART00000064554
|
ppp2r3c
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr5_+_56277866 | 8.42 |
ENSDART00000170610
ENSDART00000028854 ENSDART00000148749 |
aatf
|
apoptosis antagonizing transcription factor |
| chr3_+_25990052 | 8.42 |
ENSDART00000007398
|
gcat
|
glycine C-acetyltransferase |
| chr4_-_69787943 | 8.34 |
ENSDART00000166487
|
znf1081
|
zinc finger protein 1081 |
| chr4_+_65332578 | 8.33 |
ENSDART00000170824
|
znf1108
|
zinc finger protein 1108 |
| chr4_+_48769555 | 8.20 |
ENSDART00000150725
|
znf1020
|
zinc finger protein 1020 |
| chr10_+_23099890 | 8.18 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr4_-_28958601 | 8.13 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr4_-_32888436 | 8.04 |
ENSDART00000176728
|
BX324003.2
|
|
| chr13_+_30035253 | 8.03 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr4_+_69375564 | 8.02 |
ENSDART00000150334
|
si:dkey-246j6.1
|
si:dkey-246j6.1 |
| chr1_-_23268569 | 8.01 |
ENSDART00000143948
|
rfc1
|
replication factor C (activator 1) 1 |
| chr19_+_27339848 | 8.00 |
ENSDART00000052355
|
ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr4_+_36150332 | 8.00 |
ENSDART00000161697
|
znf1116
|
zinc finger protein 1116 |
| chr4_-_63980140 | 7.85 |
ENSDART00000160818
|
znf1091
|
zinc finger protein 1091 |
| chr8_+_471342 | 7.83 |
ENSDART00000167205
|
nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr4_-_33189070 | 7.83 |
ENSDART00000150444
|
CR759833.1
|
|
| chr4_-_40883162 | 7.82 |
ENSDART00000151978
|
znf1102
|
zinc finger protein 1102 |
| chr15_+_847077 | 7.76 |
ENSDART00000188586
|
si:dkey-7i4.5
|
si:dkey-7i4.5 |
| chr4_-_64542903 | 7.75 |
ENSDART00000143176
|
BX936382.1
|
|
| chr4_-_30712588 | 7.74 |
ENSDART00000142393
|
si:dkey-16p19.5
|
si:dkey-16p19.5 |
| chr12_-_25201576 | 7.72 |
ENSDART00000077188
|
cox7a3
|
cytochrome c oxidase subunit VIIa polypeptide 3 |
| chr12_+_36109507 | 7.65 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr4_-_43056627 | 7.64 |
ENSDART00000150269
|
si:dkey-54j5.2
|
si:dkey-54j5.2 |
| chr21_-_11199366 | 7.59 |
ENSDART00000167666
|
dnajc21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr4_-_60356042 | 7.58 |
ENSDART00000125171
|
znf1121
|
zinc finger protein 1121 |
| chr4_+_64608808 | 7.54 |
ENSDART00000143989
|
si:ch211-223a21.6
|
si:ch211-223a21.6 |
| chr1_+_29862074 | 7.51 |
ENSDART00000133905
ENSDART00000136786 ENSDART00000135252 |
pcca
|
propionyl CoA carboxylase, alpha polypeptide |
| chr15_+_841383 | 7.49 |
ENSDART00000115077
|
zgc:174573
|
zgc:174573 |
| chr4_-_32179699 | 7.47 |
ENSDART00000124106
ENSDART00000158835 |
si:dkey-72l17.6
|
si:dkey-72l17.6 |
| chr4_-_43071336 | 7.38 |
ENSDART00000150773
|
znf1072
|
zinc finger protein 1072 |
| chr5_+_54555567 | 7.36 |
ENSDART00000171159
|
anapc2
|
anaphase promoting complex subunit 2 |
| chr4_+_73651053 | 7.35 |
ENSDART00000150532
ENSDART00000172536 |
znf989
|
zinc finger protein 989 |
| chr7_-_52949944 | 7.26 |
ENSDART00000174289
|
tubgcp4
|
tubulin, gamma complex associated protein 4 |
| chr4_+_63992019 | 7.23 |
ENSDART00000158645
|
CR790365.2
|
|
| chr4_-_64482414 | 7.22 |
ENSDART00000159000
|
znf1105
|
zinc finger protein 1105 |
| chr3_-_60027255 | 7.21 |
ENSDART00000189252
ENSDART00000154684 |
recql5
|
RecQ helicase-like 5 |
| chr4_-_61920018 | 7.17 |
ENSDART00000164832
|
znf1056
|
zinc finger protein 1056 |
| chr4_-_41825115 | 7.16 |
ENSDART00000160196
|
si:dkey-237m9.1
|
si:dkey-237m9.1 |
| chr19_-_32914227 | 7.16 |
ENSDART00000186115
ENSDART00000124246 |
mtdha
|
metadherin a |
| chr4_-_64918854 | 7.15 |
ENSDART00000132409
|
si:ch211-234c11.2
|
si:ch211-234c11.2 |
| chr4_+_47385355 | 7.13 |
ENSDART00000163413
|
znf1122
|
zinc finger protein 1122 |
| chr19_+_2876106 | 7.13 |
ENSDART00000189309
|
hgh1
|
HGH1 homolog (S. cerevisiae) |
| chr11_+_12744575 | 7.11 |
ENSDART00000131059
ENSDART00000081335 ENSDART00000142481 |
rbb4l
|
retinoblastoma binding protein 4, like |
| chr4_-_43640507 | 7.10 |
ENSDART00000150700
|
si:dkey-29p23.2
|
si:dkey-29p23.2 |
| chr4_+_65271779 | 7.09 |
ENSDART00000134257
|
znf1027
|
zinc finger protein 1027 |
| chr4_+_45195202 | 6.97 |
ENSDART00000150838
ENSDART00000187011 |
znf1021
BX649479.1
|
zinc finger protein 1021 |
| chr4_-_42171503 | 6.89 |
ENSDART00000163798
|
znf1148
|
zinc finger protein 1148 |
| chr4_-_44573692 | 6.89 |
ENSDART00000153885
|
si:dkey-7j22.1
|
si:dkey-7j22.1 |
| chr5_+_1089600 | 6.83 |
ENSDART00000183128
|
LO017799.1
|
|
| chr4_+_61810270 | 6.83 |
ENSDART00000158840
|
CABZ01038376.1
|
|
| chr4_-_33405483 | 6.74 |
ENSDART00000150625
|
CT027578.1
|
|
| chr4_+_60001731 | 6.73 |
ENSDART00000150615
|
znf1109
|
zinc finger protein 1109 |
| chr4_+_42295479 | 6.72 |
ENSDART00000162279
|
znf1118
|
zinc finger protein 1118 |
| chr4_+_69409331 | 6.71 |
ENSDART00000159440
|
si:dkey-246j6.3
|
si:dkey-246j6.3 |
| chr4_+_69190524 | 6.70 |
ENSDART00000168009
|
znf1075
|
zinc finger protein 1075 |
| chr4_+_44577872 | 6.68 |
ENSDART00000157214
|
si:dkey-7j22.2
|
si:dkey-7j22.2 |
| chr4_+_68456852 | 6.66 |
ENSDART00000165623
|
znf1146
|
zinc finger protein 1146 |
| chr4_-_55178568 | 6.66 |
ENSDART00000165288
|
znf1039
|
zinc finger protein 1039 |
| chr3_+_40284598 | 6.62 |
ENSDART00000009411
|
bud31
|
BUD31 homolog (S. cerevisiae) |
| chr4_+_43679421 | 6.61 |
ENSDART00000132945
|
zgc:173705
|
zgc:173705 |
| chr4_-_53168673 | 6.58 |
ENSDART00000168891
|
znf1053
|
zinc finger protein 1053 |
| chr4_-_55322519 | 6.58 |
ENSDART00000143067
|
znf998
|
zinc finger protein 998 |
| chr19_-_26923273 | 6.57 |
ENSDART00000089609
|
skiv2l
|
SKI2 homolog, superkiller viralicidic activity 2-like |
| chr4_+_50970601 | 6.54 |
ENSDART00000150374
|
si:dkey-156j15.1
|
si:dkey-156j15.1 |
| chr4_+_38424204 | 6.47 |
ENSDART00000168884
|
znf994
|
zinc finger protein 994 |
Network of associatons between targets according to the STRING database.
First level regulatory network of creb1a+creb1b+crema+cremb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.5 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 5.0 | 14.9 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 4.5 | 13.4 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 3.9 | 19.3 | GO:0071320 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 2.9 | 14.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 2.8 | 8.4 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 2.6 | 7.8 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 2.4 | 11.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 2.2 | 9.0 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 2.1 | 6.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 2.0 | 36.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 2.0 | 6.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 1.9 | 13.6 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 1.7 | 13.2 | GO:0036372 | opsin transport(GO:0036372) |
| 1.6 | 4.9 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 1.6 | 17.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 1.5 | 20.5 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 1.4 | 4.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 1.3 | 3.9 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 1.2 | 5.0 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 1.2 | 8.7 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 1.2 | 3.6 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.0 | 7.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 1.0 | 10.0 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.9 | 8.4 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.9 | 3.7 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.9 | 5.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.8 | 19.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.8 | 3.3 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.8 | 3.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.8 | 3.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.7 | 3.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.6 | 10.9 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.6 | 10.5 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.6 | 4.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.6 | 2.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.6 | 3.3 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.5 | 4.4 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.5 | 3.8 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.5 | 17.4 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.5 | 4.9 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.4 | 50.8 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.4 | 12.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.4 | 28.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.4 | 2.6 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.4 | 4.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.4 | 1448.1 | GO:0006355 | regulation of transcription, DNA-templated(GO:0006355) |
| 0.3 | 8.0 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.3 | 3.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.3 | 10.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.3 | 2.0 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.3 | 6.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.3 | 11.1 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.3 | 13.1 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.3 | 3.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.2 | 11.8 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.2 | 1.2 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 4.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 6.5 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.2 | 3.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 2.0 | GO:0045117 | azole transport(GO:0045117) |
| 0.2 | 2.6 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.2 | 8.7 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.2 | 3.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 2.0 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.1 | 5.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 3.9 | GO:0003146 | heart jogging(GO:0003146) |
| 0.1 | 4.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 14.1 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.1 | 3.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 16.9 | GO:0006260 | DNA replication(GO:0006260) |
| 0.1 | 7.1 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.1 | 3.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 10.2 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.1 | 10.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 23.4 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 6.0 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.1 | 4.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 13.8 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 13.0 | GO:0016070 | RNA metabolic process(GO:0016070) |
| 0.0 | 3.5 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 12.2 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 7.5 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 9.7 | GO:0051726 | regulation of cell cycle(GO:0051726) |
| 0.0 | 7.4 | GO:0051301 | cell division(GO:0051301) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.8 | GO:0070545 | PeBoW complex(GO:0070545) |
| 4.5 | 13.4 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 4.3 | 17.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 4.0 | 12.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 3.1 | 9.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 2.2 | 8.9 | GO:0070209 | ASTRA complex(GO:0070209) |
| 2.0 | 10.0 | GO:0055087 | Ski complex(GO:0055087) |
| 1.7 | 19.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 1.7 | 13.6 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 1.6 | 11.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 1.5 | 16.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 1.5 | 6.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 1.4 | 9.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 1.2 | 3.7 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 1.1 | 4.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.9 | 12.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.8 | 3.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.8 | 29.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.7 | 13.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.6 | 4.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.6 | 4.9 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.6 | 5.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.5 | 6.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.5 | 2.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.5 | 7.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.5 | 4.0 | GO:0035060 | brahma complex(GO:0035060) |
| 0.5 | 10.9 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.5 | 5.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.4 | 10.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.3 | 5.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 79.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.2 | 5.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 15.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 2.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 3.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 18.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 11.8 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 6.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 2.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 879.4 | GO:0005634 | nucleus(GO:0005634) |
| 0.1 | 4.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.0 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.1 | 3.6 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 9.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 3.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 38.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 6.3 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 10.6 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 4.5 | 18.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 4.1 | 12.3 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 3.6 | 10.9 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 3.5 | 17.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 2.8 | 14.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 2.1 | 8.4 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 2.0 | 6.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 1.9 | 21.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.7 | 5.1 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 1.6 | 7.8 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 1.5 | 9.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 1.5 | 32.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 1.5 | 4.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 1.5 | 10.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.4 | 8.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 1.0 | 3.1 | GO:0000996 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 1.0 | 7.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.9 | 7.5 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.9 | 13.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.8 | 16.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.7 | 4.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.7 | 5.6 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.6 | 4.9 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.6 | 1419.3 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.5 | 10.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.5 | 8.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.5 | 2.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.5 | 11.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.5 | 5.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.4 | 35.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.4 | 4.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.3 | 23.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.3 | 3.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.3 | 5.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 2.0 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.2 | 3.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 3.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 5.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 4.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 6.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 2.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 14.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 9.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.2 | 9.3 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 3.9 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 9.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 9.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 6.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 7.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 107.5 | GO:0003723 | RNA binding(GO:0003723) |
| 0.1 | 12.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 15.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 13.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 11.9 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 1.7 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 7.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 3.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 4.6 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.7 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 4.9 | GO:0005543 | phospholipid binding(GO:0005543) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 13.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.7 | 34.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.6 | 10.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 9.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 10.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 8.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 8.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 8.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 16.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.8 | 10.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.8 | 13.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.6 | 30.3 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.6 | 10.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.6 | 13.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.5 | 4.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.5 | 7.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 10.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 4.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 7.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.4 | 12.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.3 | 6.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 10.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.2 | 5.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 8.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 4.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 11.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 5.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 3.1 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.1 | 10.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |