Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
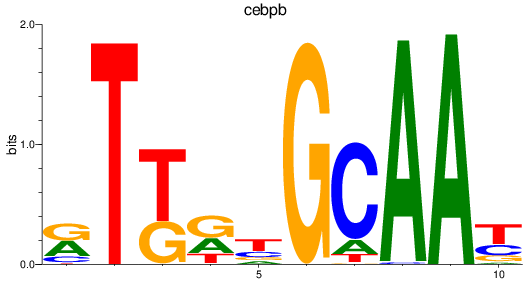
Results for cebpb
Z-value: 2.40
Transcription factors associated with cebpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpb
|
ENSDARG00000042725 | CCAAT enhancer binding protein beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpb | dr11_v1_chr8_-_28449782_28449782 | -0.77 | 1.1e-04 | Click! |
Activity profile of cebpb motif
Sorted Z-values of cebpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_8606883 | 12.68 |
ENSDART00000054469
ENSDART00000185264 |
s100a10a
|
S100 calcium binding protein A10a |
| chr5_+_51079504 | 10.84 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr6_-_345503 | 10.70 |
ENSDART00000168901
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr5_-_33259079 | 9.93 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr7_+_65240227 | 8.46 |
ENSDART00000168287
|
bco1l
|
beta-carotene oxygenase 1, like |
| chr25_+_31222069 | 8.37 |
ENSDART00000159373
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr2_-_32643738 | 8.02 |
ENSDART00000112452
|
si:dkeyp-73d8.9
|
si:dkeyp-73d8.9 |
| chr6_-_43449013 | 7.77 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr12_+_46960579 | 7.75 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr20_-_40755614 | 6.92 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr25_-_30429607 | 6.83 |
ENSDART00000162429
ENSDART00000176535 |
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr21_-_40174647 | 6.67 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr5_-_30615901 | 6.64 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr2_-_30460293 | 6.62 |
ENSDART00000113193
|
cbln2a
|
cerebellin 2a precursor |
| chr5_-_58840971 | 6.38 |
ENSDART00000050932
|
tmem136b
|
transmembrane protein 136b |
| chr9_-_48736388 | 6.13 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr24_-_40009446 | 6.08 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr2_-_5356686 | 6.03 |
ENSDART00000124290
|
MFN1
|
mitofusin 1 |
| chr1_-_9231952 | 5.87 |
ENSDART00000166515
|
si:dkeyp-57d7.4
|
si:dkeyp-57d7.4 |
| chr16_+_25245857 | 5.82 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr7_+_49862837 | 5.79 |
ENSDART00000174315
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr9_-_34915351 | 5.78 |
ENSDART00000100728
ENSDART00000139608 |
upf3a
|
UPF3A, regulator of nonsense mediated mRNA decay |
| chr7_-_34339845 | 5.54 |
ENSDART00000173816
|
madd
|
MAP-kinase activating death domain |
| chr10_-_17103651 | 5.46 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr22_+_17203752 | 5.44 |
ENSDART00000143376
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr23_-_26521970 | 5.38 |
ENSDART00000143712
|
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr14_-_46238186 | 5.37 |
ENSDART00000173245
|
si:ch211-113d11.6
|
si:ch211-113d11.6 |
| chr21_-_43550120 | 5.27 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr14_+_16345003 | 5.16 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr24_-_39771667 | 5.11 |
ENSDART00000181867
|
GFOD1
|
si:ch211-276f18.2 |
| chr3_+_19299309 | 5.05 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr4_-_9549693 | 4.92 |
ENSDART00000160242
|
FQ377934.1
|
|
| chr8_-_39739627 | 4.86 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr9_-_9989660 | 4.86 |
ENSDART00000081463
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr20_+_1121458 | 4.81 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr4_+_12113105 | 4.72 |
ENSDART00000182399
|
tmem178b
|
transmembrane protein 178B |
| chr21_-_4032650 | 4.69 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr12_+_35119762 | 4.68 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr2_-_31661609 | 4.63 |
ENSDART00000144383
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr15_-_1885247 | 4.60 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr17_+_30450163 | 4.46 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr7_-_7845540 | 4.36 |
ENSDART00000166280
|
cxcl8b.1
|
chemokine (C-X-C motif) ligand 8b, duplicate 1 |
| chr7_-_7823662 | 4.35 |
ENSDART00000167652
|
cxcl8b.3
|
chemokine (C-X-C motif) ligand 8b, duplicate 3 |
| chr12_-_14922955 | 4.35 |
ENSDART00000002078
|
neurod2
|
neurogenic differentiation 2 |
| chr9_-_53062083 | 4.21 |
ENSDART00000122155
|
zmp:0000000936
|
zmp:0000000936 |
| chr16_-_27749172 | 4.19 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr5_+_26795773 | 4.12 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr23_-_18030399 | 3.99 |
ENSDART00000136967
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr17_-_14613711 | 3.94 |
ENSDART00000157345
|
sdsl
|
serine dehydratase-like |
| chr18_-_20869175 | 3.91 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr13_+_40437550 | 3.86 |
ENSDART00000057090
ENSDART00000167859 |
got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr20_+_43113465 | 3.83 |
ENSDART00000004842
|
dusp23a
|
dual specificity phosphatase 23a |
| chr14_-_33613794 | 3.82 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr4_-_14915268 | 3.77 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr19_+_1370504 | 3.76 |
ENSDART00000158946
|
dgat1a
|
diacylglycerol O-acyltransferase 1a |
| chr17_+_30587333 | 3.74 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr19_-_9829965 | 3.71 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr25_-_19443421 | 3.66 |
ENSDART00000067362
|
cart2
|
cocaine- and amphetamine-regulated transcript 2 |
| chr19_+_5674907 | 3.65 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr15_-_12258851 | 3.59 |
ENSDART00000180656
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr21_-_43949208 | 3.56 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr8_+_25351863 | 3.55 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr14_-_12071447 | 3.50 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr5_-_69948099 | 3.45 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr14_+_36885524 | 3.40 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr9_+_219124 | 3.37 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr17_+_3124129 | 3.36 |
ENSDART00000155323
|
zgc:136872
|
zgc:136872 |
| chr6_+_8314451 | 3.34 |
ENSDART00000147793
ENSDART00000183688 |
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr11_-_6265574 | 3.32 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr13_-_20540790 | 3.32 |
ENSDART00000131467
|
si:ch1073-126c3.2
|
si:ch1073-126c3.2 |
| chr20_-_25533739 | 3.32 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr7_+_4162994 | 3.30 |
ENSDART00000172800
|
si:ch211-63p21.1
|
si:ch211-63p21.1 |
| chr2_+_31476065 | 3.29 |
ENSDART00000049219
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr12_-_33706726 | 3.20 |
ENSDART00000153135
|
myo15b
|
myosin XVB |
| chr25_+_20188769 | 3.19 |
ENSDART00000142781
|
cald1b
|
caldesmon 1b |
| chr9_-_1702648 | 3.19 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr1_-_19237576 | 3.17 |
ENSDART00000143663
ENSDART00000131579 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr18_-_16179129 | 3.17 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr14_-_5642371 | 3.16 |
ENSDART00000183859
ENSDART00000054876 |
npm1b
|
nucleophosmin 1b |
| chr11_-_29082175 | 3.14 |
ENSDART00000123245
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr24_-_38644937 | 3.12 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr2_-_48966431 | 3.10 |
ENSDART00000147948
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr18_+_15644559 | 3.10 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr17_-_45552602 | 3.08 |
ENSDART00000154844
ENSDART00000034432 |
susd4
|
sushi domain containing 4 |
| chr12_-_45304971 | 3.06 |
ENSDART00000186537
ENSDART00000126405 |
fdxr
|
ferredoxin reductase |
| chr13_+_7292061 | 3.02 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr21_-_5007109 | 3.02 |
ENSDART00000187042
ENSDART00000097796 ENSDART00000146766 |
rnf165a
|
ring finger protein 165a |
| chr11_-_2250767 | 3.01 |
ENSDART00000018131
|
hnrnpa1a
|
heterogeneous nuclear ribonucleoprotein A1a |
| chr1_+_50538839 | 2.99 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr24_-_31123365 | 2.98 |
ENSDART00000182947
|
tmem56a
|
transmembrane protein 56a |
| chr18_-_14274803 | 2.95 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr25_-_32869794 | 2.93 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr22_-_10487490 | 2.89 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr10_+_44940693 | 2.87 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr12_-_31012741 | 2.85 |
ENSDART00000145967
|
tcf7l2
|
transcription factor 7 like 2 |
| chr19_+_4990496 | 2.85 |
ENSDART00000151050
ENSDART00000017535 |
zgc:91968
|
zgc:91968 |
| chr7_+_11543999 | 2.79 |
ENSDART00000173676
|
il16
|
interleukin 16 |
| chr9_+_13682133 | 2.78 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr16_+_49601838 | 2.77 |
ENSDART00000168570
ENSDART00000159236 |
si:dkey-82o10.4
|
si:dkey-82o10.4 |
| chr22_+_18469004 | 2.74 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr14_-_7306983 | 2.73 |
ENSDART00000158914
|
si:ch211-51f19.1
|
si:ch211-51f19.1 |
| chr23_-_36724575 | 2.72 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr16_+_25259313 | 2.72 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr16_+_47207691 | 2.72 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr17_-_15149192 | 2.71 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr1_-_19215336 | 2.70 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr7_-_4296771 | 2.65 |
ENSDART00000128855
|
cbln11
|
cerebellin 11 |
| chr15_-_4314042 | 2.64 |
ENSDART00000173311
ENSDART00000170562 |
ECE2
|
si:ch211-117a13.2 |
| chr22_-_18491813 | 2.64 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr13_+_13770980 | 2.61 |
ENSDART00000113089
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr14_+_7902374 | 2.61 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr7_-_29723761 | 2.60 |
ENSDART00000173560
|
vps13c
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr16_+_50289916 | 2.57 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr17_-_6076084 | 2.56 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr22_-_16377666 | 2.55 |
ENSDART00000161878
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr6_+_27151940 | 2.55 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr16_+_14029283 | 2.55 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr25_+_13191391 | 2.55 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr24_-_28381404 | 2.50 |
ENSDART00000148406
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr21_-_39058490 | 2.49 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr11_-_24681292 | 2.48 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr3_+_13637383 | 2.47 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr15_-_22139566 | 2.45 |
ENSDART00000149017
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr15_+_45994123 | 2.44 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr23_-_36003441 | 2.43 |
ENSDART00000164699
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr6_-_38816500 | 2.42 |
ENSDART00000190866
ENSDART00000104124 |
cnga3a
|
cyclic nucleotide gated channel alpha 3a |
| chr19_-_9662958 | 2.42 |
ENSDART00000041094
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr12_-_31457301 | 2.42 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr12_+_16132612 | 2.41 |
ENSDART00000152550
|
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr25_-_17239008 | 2.41 |
ENSDART00000151965
ENSDART00000152106 ENSDART00000152107 |
ccnd2a
|
cyclin D2, a |
| chr23_+_44732863 | 2.40 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr19_+_7045033 | 2.37 |
ENSDART00000146579
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr20_+_16743056 | 2.33 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr2_+_11029138 | 2.32 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr25_-_32311048 | 2.30 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr19_+_4990320 | 2.29 |
ENSDART00000147056
|
zgc:91968
|
zgc:91968 |
| chr18_+_507835 | 2.25 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr4_-_77432218 | 2.23 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr10_+_15255012 | 2.21 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr8_+_28900689 | 2.20 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr3_-_57744323 | 2.19 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr9_-_4856767 | 2.16 |
ENSDART00000016814
ENSDART00000138015 |
fmnl2a
|
formin-like 2a |
| chr19_-_9472893 | 2.14 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr6_+_8315050 | 2.12 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr6_-_14292307 | 2.08 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr11_-_44931962 | 2.07 |
ENSDART00000170345
|
pfklb
|
phosphofructokinase, liver b |
| chr24_-_15648636 | 2.05 |
ENSDART00000136200
|
cbln2b
|
cerebellin 2b precursor |
| chr17_-_6076266 | 2.04 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr3_-_43821191 | 2.03 |
ENSDART00000186816
|
snn
|
stannin |
| chr5_+_32162684 | 2.02 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr25_+_13191615 | 2.01 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr8_+_46386601 | 2.01 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr3_+_22242269 | 2.00 |
ENSDART00000168970
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr22_-_26175237 | 2.00 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr18_+_22287084 | 1.98 |
ENSDART00000151919
ENSDART00000181644 |
ctcf
|
CCCTC-binding factor (zinc finger protein) |
| chr21_-_36972127 | 1.97 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr19_+_22085925 | 1.96 |
ENSDART00000185636
|
atp9b
|
ATPase phospholipid transporting 9B |
| chr1_+_14253118 | 1.96 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr10_+_15310811 | 1.95 |
ENSDART00000136239
|
kcnv2a
|
potassium channel, subfamily V, member 2a |
| chr12_+_32073660 | 1.94 |
ENSDART00000153245
ENSDART00000153268 |
stxbp4
|
syntaxin binding protein 4 |
| chr9_+_33154841 | 1.94 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr22_+_15438513 | 1.94 |
ENSDART00000010846
|
gpc5b
|
glypican 5b |
| chr23_-_24195723 | 1.93 |
ENSDART00000145489
|
ano11
|
anoctamin 11 |
| chr17_+_443264 | 1.92 |
ENSDART00000159086
|
zgc:195050
|
zgc:195050 |
| chr23_+_2669 | 1.89 |
ENSDART00000011146
|
twist3
|
twist3 |
| chr6_-_13498745 | 1.87 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr2_-_36819624 | 1.87 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr24_+_26134029 | 1.87 |
ENSDART00000185134
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr8_+_16990120 | 1.85 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr1_+_55752593 | 1.84 |
ENSDART00000108838
ENSDART00000134770 |
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr10_+_15255198 | 1.83 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr23_-_33361425 | 1.81 |
ENSDART00000031638
|
slc48a1a
|
solute carrier family 48 (heme transporter), member 1a |
| chr16_+_44298902 | 1.81 |
ENSDART00000114795
|
dpys
|
dihydropyrimidinase |
| chr5_+_8196264 | 1.81 |
ENSDART00000174564
ENSDART00000161261 |
lmbrd2a
|
LMBR1 domain containing 2a |
| chr12_-_46959990 | 1.80 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr7_+_16033923 | 1.80 |
ENSDART00000161669
ENSDART00000114062 |
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr10_+_45248326 | 1.79 |
ENSDART00000159797
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr13_-_31544365 | 1.79 |
ENSDART00000005670
|
dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr14_-_2322484 | 1.78 |
ENSDART00000167806
|
si:ch73-379j16.2
|
si:ch73-379j16.2 |
| chr7_-_52842605 | 1.77 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr22_+_15438872 | 1.77 |
ENSDART00000139800
|
gpc5b
|
glypican 5b |
| chr17_+_10501647 | 1.76 |
ENSDART00000140391
|
tyro3
|
TYRO3 protein tyrosine kinase |
| chr15_-_29573267 | 1.76 |
ENSDART00000099947
|
samsn1a
|
SAM domain, SH3 domain and nuclear localisation signals 1a |
| chr21_-_22547496 | 1.75 |
ENSDART00000166835
ENSDART00000089030 |
myo5b
|
myosin VB |
| chr5_-_55395384 | 1.75 |
ENSDART00000147298
ENSDART00000082577 |
prune2
|
prune homolog 2 (Drosophila) |
| chr17_-_43466317 | 1.74 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr20_+_9211237 | 1.72 |
ENSDART00000139527
|
si:ch211-59d15.4
|
si:ch211-59d15.4 |
| chr1_-_52222989 | 1.71 |
ENSDART00000010236
|
cnn1a
|
calponin 1, basic, smooth muscle, a |
| chr5_-_26181863 | 1.69 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr17_-_27976031 | 1.68 |
ENSDART00000154829
|
ptafr
|
platelet-activating factor receptor |
| chr22_+_26798853 | 1.65 |
ENSDART00000087576
ENSDART00000179780 |
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr22_-_26236188 | 1.64 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr19_-_15192840 | 1.61 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr25_+_13498188 | 1.59 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr5_-_56412262 | 1.58 |
ENSDART00000083079
|
acaca
|
acetyl-CoA carboxylase alpha |
| chr24_-_17049270 | 1.58 |
ENSDART00000175508
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr11_+_37251825 | 1.57 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr6_+_22068589 | 1.55 |
ENSDART00000151205
|
aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr5_+_19094153 | 1.54 |
ENSDART00000186525
ENSDART00000064752 |
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr6_+_58915889 | 1.54 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 10.7 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 1.4 | 4.1 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 1.2 | 6.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 1.1 | 3.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 1.1 | 4.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 1.1 | 6.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 1.0 | 4.2 | GO:0015677 | copper ion import(GO:0015677) |
| 1.0 | 3.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 1.0 | 4.0 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 1.0 | 3.0 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 1.0 | 3.9 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 1.0 | 3.9 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.9 | 8.5 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.9 | 8.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.9 | 4.5 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.8 | 5.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.8 | 2.4 | GO:2000193 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) positive regulation of lipid transport(GO:0032370) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.8 | 3.2 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.7 | 2.9 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.7 | 5.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.7 | 2.7 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.6 | 2.5 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.6 | 5.8 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.6 | 2.8 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.5 | 1.5 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.5 | 2.9 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.5 | 2.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.5 | 6.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 1.4 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.4 | 1.7 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.4 | 1.7 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.4 | 1.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.4 | 2.6 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.4 | 5.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 2.6 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.4 | 3.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 4.4 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.3 | 3.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.3 | 1.3 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.3 | 5.1 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.3 | 4.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.3 | 1.4 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.3 | 2.8 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.3 | 2.6 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.3 | 2.9 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.3 | 1.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.3 | 1.0 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.2 | 8.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.2 | 1.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 3.7 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.2 | 3.6 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.2 | 1.8 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.2 | 3.1 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 2.1 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.8 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 0.9 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 1.0 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.2 | 4.6 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.2 | 4.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 2.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.2 | 5.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 1.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 8.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 3.1 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 2.4 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.2 | 1.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 1.7 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 0.5 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 2.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 5.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 2.2 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.2 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 2.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 2.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.8 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 2.4 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 1.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 2.7 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 2.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 1.2 | GO:0098869 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 2.9 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 1.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 1.0 | GO:0042698 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.1 | 0.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 5.2 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 3.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 3.0 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.6 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 0.8 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 2.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 3.6 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 0.5 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.1 | 0.3 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 2.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 1.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 4.5 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 2.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 3.2 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.1 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.9 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 1.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 1.8 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 4.8 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.1 | 1.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 1.8 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 3.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 3.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 4.0 | GO:1902652 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
| 0.0 | 0.6 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 1.0 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.7 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 2.5 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.4 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 2.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.6 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 2.1 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 0.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 3.6 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 1.5 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 1.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 2.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.9 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 1.6 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.6 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 1.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.9 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 3.2 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.8 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 2.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.2 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 3.4 | GO:0009725 | response to hormone(GO:0009725) |
| 0.0 | 0.6 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.5 | 10.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.7 | 2.9 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.5 | 2.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 5.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 1.8 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.3 | 1.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.3 | 3.9 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.3 | 1.7 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.2 | 2.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 4.7 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 2.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 3.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 8.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 4.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 7.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 1.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 3.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.5 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 2.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 4.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 9.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 2.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 3.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 3.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 3.9 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 3.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 27.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 13.6 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.1 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.1 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 1.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 13.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.7 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 2.3 | 9.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 1.5 | 4.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 1.4 | 5.5 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 1.2 | 6.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.1 | 4.6 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 1.0 | 4.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.0 | 3.9 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.9 | 8.5 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.9 | 7.8 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.8 | 4.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.8 | 10.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.8 | 3.9 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.8 | 6.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.7 | 2.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.6 | 2.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.6 | 3.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.6 | 2.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.6 | 6.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.5 | 3.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.5 | 4.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.5 | 1.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.5 | 2.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.5 | 3.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 12.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.5 | 1.8 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.4 | 2.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.4 | 1.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 1.6 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.4 | 2.6 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.4 | 3.8 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.4 | 3.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 1.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.4 | 4.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 3.6 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 3.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 2.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 7.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.3 | 2.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 2.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 3.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 1.0 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.3 | 1.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 1.0 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 2.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 5.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 5.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 0.9 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 0.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 2.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 5.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.2 | 2.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 2.4 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 3.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 1.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 1.7 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 1.8 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 2.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 8.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 2.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 2.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 4.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 4.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.4 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.1 | 2.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 3.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.7 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 3.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 3.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.4 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 0.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.6 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 3.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.4 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.2 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.1 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 3.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.5 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 2.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 4.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 0.8 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 2.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 3.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 2.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 2.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 8.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.9 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.9 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 2.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 3.2 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 9.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 3.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 4.9 | GO:0022832 | voltage-gated channel activity(GO:0022832) |
| 0.0 | 1.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 4.0 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 1.5 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 5.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 2.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 3.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 4.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 5.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 3.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 4.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 12.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.2 | 3.6 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.8 | 6.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.6 | 10.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.4 | 3.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 2.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 4.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.3 | 2.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 1.6 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.2 | 1.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 3.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 1.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 2.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 0.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 3.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 2.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 4.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.0 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 4.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 2.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.0 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |