Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
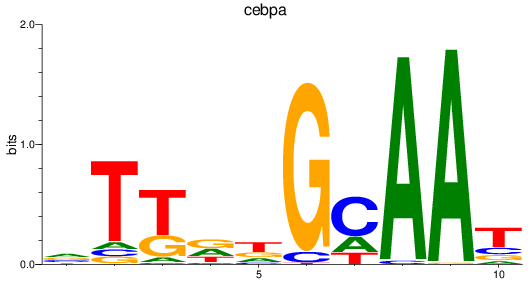
Results for cebpa
Z-value: 0.78
Transcription factors associated with cebpa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpa
|
ENSDARG00000036074 | CCAAT enhancer binding protein alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpa | dr11_v1_chr7_-_38087865_38087865 | -0.17 | 5.0e-01 | Click! |
Activity profile of cebpa motif
Sorted Z-values of cebpa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_8606883 | 1.81 |
ENSDART00000054469
ENSDART00000185264 |
s100a10a
|
S100 calcium binding protein A10a |
| chr25_+_31222069 | 1.35 |
ENSDART00000159373
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr6_-_43449013 | 1.24 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr2_-_51794472 | 1.14 |
ENSDART00000186652
|
BX908782.3
|
|
| chr5_+_45677781 | 1.05 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr6_-_345503 | 0.99 |
ENSDART00000168901
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr1_+_9954489 | 0.86 |
ENSDART00000005895
ENSDART00000183003 |
pdia2
|
protein disulfide isomerase family A, member 2 |
| chr6_+_41186320 | 0.80 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr7_+_19903924 | 0.79 |
ENSDART00000159112
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr16_+_50289916 | 0.79 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr19_+_5674907 | 0.76 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr14_+_15155684 | 0.75 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr7_+_19904136 | 0.72 |
ENSDART00000173452
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr4_-_9549693 | 0.69 |
ENSDART00000160242
|
FQ377934.1
|
|
| chr7_-_7845540 | 0.69 |
ENSDART00000166280
|
cxcl8b.1
|
chemokine (C-X-C motif) ligand 8b, duplicate 1 |
| chr2_-_32643738 | 0.68 |
ENSDART00000112452
|
si:dkeyp-73d8.9
|
si:dkeyp-73d8.9 |
| chr7_+_22616212 | 0.66 |
ENSDART00000052844
|
cldn7a
|
claudin 7a |
| chr9_+_32301017 | 0.65 |
ENSDART00000127916
ENSDART00000183298 ENSDART00000143103 |
hspe1
|
heat shock 10 protein 1 |
| chr2_-_30460293 | 0.60 |
ENSDART00000113193
|
cbln2a
|
cerebellin 2a precursor |
| chr8_-_13972626 | 0.59 |
ENSDART00000144296
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr24_-_36238054 | 0.59 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr15_-_12258851 | 0.58 |
ENSDART00000180656
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr1_+_34203817 | 0.56 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr21_-_22709251 | 0.56 |
ENSDART00000140032
|
si:dkeyp-69c1.9
|
si:dkeyp-69c1.9 |
| chr14_+_36885524 | 0.55 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr5_-_58840971 | 0.54 |
ENSDART00000050932
|
tmem136b
|
transmembrane protein 136b |
| chr7_+_42935126 | 0.53 |
ENSDART00000157747
|
BX284696.1
|
|
| chr9_-_48736388 | 0.52 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr12_-_32073611 | 0.51 |
ENSDART00000153369
ENSDART00000086531 |
cox11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr23_+_29889089 | 0.51 |
ENSDART00000149378
ENSDART00000089465 |
mxra8b
|
matrix-remodelling associated 8b |
| chr13_-_20540790 | 0.50 |
ENSDART00000131467
|
si:ch1073-126c3.2
|
si:ch1073-126c3.2 |
| chr18_-_40905901 | 0.48 |
ENSDART00000064848
|
pglyrp5
|
peptidoglycan recognition protein 5 |
| chr17_+_12942634 | 0.48 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr13_+_33688474 | 0.47 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr14_+_16345003 | 0.47 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr3_-_13068189 | 0.47 |
ENSDART00000167180
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr5_+_4332220 | 0.46 |
ENSDART00000051699
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr1_-_22757145 | 0.46 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr11_-_7156620 | 0.46 |
ENSDART00000172823
ENSDART00000172879 ENSDART00000078916 |
smim7
|
small integral membrane protein 7 |
| chr14_+_21107032 | 0.45 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr8_-_39739627 | 0.45 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr7_-_4296771 | 0.45 |
ENSDART00000128855
|
cbln11
|
cerebellin 11 |
| chr8_-_50147948 | 0.45 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr10_-_17103651 | 0.45 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr14_+_31657412 | 0.45 |
ENSDART00000105767
|
fhl1a
|
four and a half LIM domains 1a |
| chr3_-_40955780 | 0.45 |
ENSDART00000130130
|
cyp3c3
|
cytochrome P450, family 3, subfamily c, polypeptide 3 |
| chr24_-_32582378 | 0.44 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr13_-_23756700 | 0.44 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr3_-_58205045 | 0.43 |
ENSDART00000130740
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr9_-_9989660 | 0.42 |
ENSDART00000081463
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr22_-_24248420 | 0.42 |
ENSDART00000165433
|
rgs2
|
regulator of G protein signaling 2 |
| chr16_-_4640539 | 0.41 |
ENSDART00000076955
ENSDART00000131949 |
cyp4t8
|
cytochrome P450, family 4, subfamily T, polypeptide 8 |
| chr5_+_44805269 | 0.41 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr1_+_59067978 | 0.41 |
ENSDART00000172613
|
MFAP4 (1 of many)
|
si:ch1073-110a20.7 |
| chr21_+_38002879 | 0.41 |
ENSDART00000065183
|
cldn2
|
claudin 2 |
| chr19_-_40192249 | 0.40 |
ENSDART00000051972
|
grn1
|
granulin 1 |
| chr9_-_23850978 | 0.40 |
ENSDART00000132915
|
serpine3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr9_+_51265283 | 0.40 |
ENSDART00000137426
|
gcgb
|
glucagon b |
| chr21_+_20901505 | 0.40 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr12_-_35949936 | 0.39 |
ENSDART00000192583
|
AL954682.1
|
|
| chr17_-_14613711 | 0.39 |
ENSDART00000157345
|
sdsl
|
serine dehydratase-like |
| chr7_-_10560964 | 0.39 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr15_+_28202170 | 0.38 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr24_-_38094074 | 0.38 |
ENSDART00000140802
ENSDART00000137734 |
crp2
|
C-reactive protein 2 |
| chr13_+_24396666 | 0.38 |
ENSDART00000139197
ENSDART00000101200 |
zgc:153169
|
zgc:153169 |
| chr24_-_40009446 | 0.37 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr17_+_12942021 | 0.37 |
ENSDART00000192514
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr24_-_32582880 | 0.36 |
ENSDART00000186307
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr24_+_42948 | 0.36 |
ENSDART00000122785
|
tmx3b
|
thioredoxin related transmembrane protein 3b |
| chr13_+_25761279 | 0.36 |
ENSDART00000177818
ENSDART00000002863 |
neurog3
|
neurogenin 3 |
| chr12_+_17504559 | 0.35 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr10_-_36808348 | 0.35 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr16_-_38333976 | 0.35 |
ENSDART00000031895
|
cdc42se1
|
CDC42 small effector 1 |
| chr12_-_15620090 | 0.35 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr3_-_18410968 | 0.35 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr11_+_3281899 | 0.35 |
ENSDART00000181012
|
mmp19
|
matrix metallopeptidase 19 |
| chr10_-_44508249 | 0.35 |
ENSDART00000160018
|
DUSP26
|
dual specificity phosphatase 26 |
| chr5_+_44805028 | 0.35 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr16_+_23984179 | 0.35 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr10_-_27009413 | 0.34 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr7_-_7823662 | 0.34 |
ENSDART00000167652
|
cxcl8b.3
|
chemokine (C-X-C motif) ligand 8b, duplicate 3 |
| chr3_-_18805225 | 0.34 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr23_-_26535875 | 0.33 |
ENSDART00000135988
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr6_+_8314451 | 0.33 |
ENSDART00000147793
ENSDART00000183688 |
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr25_-_10564721 | 0.33 |
ENSDART00000154776
|
galn
|
galanin/GMAP prepropeptide |
| chr10_-_24343507 | 0.33 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr4_+_842010 | 0.33 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr14_+_21106444 | 0.32 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr10_-_29733194 | 0.31 |
ENSDART00000149252
|
si:ch73-261i21.2
|
si:ch73-261i21.2 |
| chr5_+_9360394 | 0.31 |
ENSDART00000124642
|
FP236810.2
|
|
| chr19_-_44054930 | 0.31 |
ENSDART00000151084
ENSDART00000150991 ENSDART00000005191 |
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr6_-_49510553 | 0.31 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr19_+_21818460 | 0.30 |
ENSDART00000189427
ENSDART00000180525 |
CU856623.1
|
|
| chr11_-_6265574 | 0.30 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr21_+_5508167 | 0.30 |
ENSDART00000157702
|
ly6m4
|
lymphocyte antigen 6 family member M4 |
| chr12_-_33706726 | 0.30 |
ENSDART00000153135
|
myo15b
|
myosin XVB |
| chr7_-_6362460 | 0.30 |
ENSDART00000129239
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr23_-_26536055 | 0.30 |
ENSDART00000182719
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr15_-_24960730 | 0.30 |
ENSDART00000109990
ENSDART00000186706 |
abhd15a
|
abhydrolase domain containing 15a |
| chr25_-_19443421 | 0.29 |
ENSDART00000067362
|
cart2
|
cocaine- and amphetamine-regulated transcript 2 |
| chr17_+_14425219 | 0.29 |
ENSDART00000153994
|
ccdc175
|
coiled-coil domain containing 175 |
| chr5_+_26795773 | 0.29 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr23_-_31633201 | 0.29 |
ENSDART00000143335
ENSDART00000053531 |
slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr24_-_27436319 | 0.29 |
ENSDART00000171489
|
si:dkey-25o1.7
|
si:dkey-25o1.7 |
| chr19_-_40191358 | 0.29 |
ENSDART00000183919
|
grn1
|
granulin 1 |
| chr5_-_30615901 | 0.29 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr21_-_29117670 | 0.29 |
ENSDART00000124327
|
havcr2
|
hepatitis A virus cellular receptor 2 |
| chr8_+_30699429 | 0.29 |
ENSDART00000005345
|
upb1
|
ureidopropionase, beta |
| chr17_-_125091 | 0.29 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr24_-_25184553 | 0.29 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr25_+_13191615 | 0.28 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr17_+_30450163 | 0.28 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr24_+_15020402 | 0.28 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr14_-_5642371 | 0.27 |
ENSDART00000183859
ENSDART00000054876 |
npm1b
|
nucleophosmin 1b |
| chr22_-_18491813 | 0.27 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr21_+_41837610 | 0.27 |
ENSDART00000157974
|
rnf14
|
ring finger protein 14 |
| chr25_-_16146851 | 0.27 |
ENSDART00000104043
|
dkk3b
|
dickkopf WNT signaling pathway inhibitor 3b |
| chr23_-_26128593 | 0.27 |
ENSDART00000136855
ENSDART00000193700 ENSDART00000163984 |
rca2.2
|
regulator of complement activation group 2 gene 2 |
| chr19_-_41213718 | 0.26 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr16_+_16977786 | 0.26 |
ENSDART00000043173
ENSDART00000132150 |
rpl18
|
ribosomal protein L18 |
| chr13_-_23270576 | 0.26 |
ENSDART00000132828
|
si:dkey-103j14.5
|
si:dkey-103j14.5 |
| chr8_-_2434282 | 0.26 |
ENSDART00000137262
ENSDART00000134044 |
vdac3
|
voltage-dependent anion channel 3 |
| chr21_-_40630650 | 0.26 |
ENSDART00000172706
|
pcyt1bb
|
phosphate cytidylyltransferase 1, choline, beta b |
| chr10_+_2234283 | 0.26 |
ENSDART00000136363
|
cntnap3
|
contactin associated protein like 3 |
| chr1_-_59130695 | 0.26 |
ENSDART00000152560
|
FP015850.1
|
|
| chr8_+_7801060 | 0.26 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr1_-_43948826 | 0.25 |
ENSDART00000132038
|
si:dkey-22i16.7
|
si:dkey-22i16.7 |
| chr5_+_8919698 | 0.25 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr4_+_76735113 | 0.25 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr11_+_37178271 | 0.24 |
ENSDART00000161771
|
itih3b
|
inter-alpha-trypsin inhibitor heavy chain 3b |
| chr10_+_6496185 | 0.24 |
ENSDART00000164770
|
reep5
|
receptor accessory protein 5 |
| chr21_-_40173821 | 0.24 |
ENSDART00000180667
|
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr21_+_40195452 | 0.24 |
ENSDART00000108650
|
or115-15
|
odorant receptor, family F, subfamily 115, member 15 |
| chr11_-_13152215 | 0.24 |
ENSDART00000160989
ENSDART00000158239 |
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr9_+_29520696 | 0.24 |
ENSDART00000144430
|
fdx1
|
ferredoxin 1 |
| chr8_+_25351863 | 0.23 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr15_+_14856307 | 0.23 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr25_+_14109626 | 0.23 |
ENSDART00000109883
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr11_-_40147032 | 0.23 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr21_+_43178831 | 0.23 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr2_-_15033136 | 0.23 |
ENSDART00000145974
ENSDART00000034865 |
hccsa.1
|
holocytochrome c synthase a |
| chr14_-_41535822 | 0.23 |
ENSDART00000149407
|
itga6l
|
integrin, alpha 6, like |
| chr25_+_20188769 | 0.23 |
ENSDART00000142781
|
cald1b
|
caldesmon 1b |
| chr16_-_21785261 | 0.23 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr1_+_34224360 | 0.23 |
ENSDART00000192938
|
arl6
|
ADP-ribosylation factor-like 6 |
| chr21_-_4032650 | 0.23 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr1_+_31110817 | 0.22 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr17_+_5623514 | 0.22 |
ENSDART00000171220
ENSDART00000176083 |
CU571310.1
|
|
| chr20_+_1121458 | 0.22 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr12_+_35046704 | 0.22 |
ENSDART00000105523
ENSDART00000149946 |
timm23a
|
translocase of inner mitochondrial membrane 23 homolog a (yeast) |
| chr5_-_3960161 | 0.22 |
ENSDART00000111453
|
myo19
|
myosin XIX |
| chr3_+_52806347 | 0.22 |
ENSDART00000168151
ENSDART00000098826 |
lmx1al
|
LIM homeobox transcription factor 1, alpha-like |
| chr13_-_9598320 | 0.22 |
ENSDART00000184613
|
cpxm1a
|
carboxypeptidase X (M14 family), member 1a |
| chr2_+_243778 | 0.22 |
ENSDART00000182262
|
CABZ01085887.1
|
|
| chr19_-_5699703 | 0.22 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr3_-_26190804 | 0.22 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr13_+_40019001 | 0.21 |
ENSDART00000158820
|
golga7bb
|
golgin A7 family, member Bb |
| chr3_-_57666518 | 0.21 |
ENSDART00000102062
|
timp2b
|
TIMP metallopeptidase inhibitor 2b |
| chr10_-_16470648 | 0.21 |
ENSDART00000149104
|
fbn2a
|
fibrillin 2a |
| chr4_-_14915268 | 0.21 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr13_+_22964868 | 0.21 |
ENSDART00000142129
|
tacr2
|
tachykinin receptor 2 |
| chr23_+_45282858 | 0.21 |
ENSDART00000162353
|
CABZ01073265.1
|
|
| chr3_-_15444396 | 0.21 |
ENSDART00000104361
|
si:dkey-56d12.4
|
si:dkey-56d12.4 |
| chr14_-_9281232 | 0.21 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr14_-_17576391 | 0.20 |
ENSDART00000161355
ENSDART00000168959 |
rnf4
|
ring finger protein 4 |
| chr9_-_21838045 | 0.20 |
ENSDART00000147471
|
acod1
|
aconitate decarboxylase 1 |
| chr7_-_73851280 | 0.20 |
ENSDART00000190053
|
FP236812.3
|
|
| chr10_+_15088534 | 0.20 |
ENSDART00000142865
|
si:ch211-95j8.3
|
si:ch211-95j8.3 |
| chr12_+_15008582 | 0.20 |
ENSDART00000003847
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr1_+_38818268 | 0.20 |
ENSDART00000166864
|
spcs3
|
signal peptidase complex subunit 3 |
| chr22_-_26236188 | 0.20 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr3_+_19299309 | 0.20 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr15_-_1885247 | 0.20 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr12_-_4370585 | 0.20 |
ENSDART00000129502
|
CA4 (1 of many)
|
si:ch211-173d10.4 |
| chr7_+_59677273 | 0.19 |
ENSDART00000039535
ENSDART00000132044 |
trmt44
|
tRNA methyltransferase 44 homolog |
| chr24_-_25788841 | 0.19 |
ENSDART00000132235
|
klhl24b
|
kelch-like family member 24b |
| chr20_-_30900947 | 0.19 |
ENSDART00000153419
ENSDART00000062536 |
hebp2
|
heme binding protein 2 |
| chr9_+_32301456 | 0.19 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr17_-_31695217 | 0.19 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr13_+_31545530 | 0.19 |
ENSDART00000164590
ENSDART00000178460 ENSDART00000185503 |
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr5_+_51079504 | 0.19 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr24_-_38644937 | 0.19 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr8_+_13503377 | 0.19 |
ENSDART00000034740
ENSDART00000167187 |
fut9d
|
fucosyltransferase 9d |
| chr22_-_36875264 | 0.19 |
ENSDART00000137548
|
kng1
|
kininogen 1 |
| chr2_-_5356686 | 0.19 |
ENSDART00000124290
|
MFN1
|
mitofusin 1 |
| chr6_-_24103666 | 0.19 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr19_-_15192840 | 0.19 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr7_-_66864756 | 0.19 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr5_-_38342992 | 0.19 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr15_-_30832897 | 0.19 |
ENSDART00000152330
|
msi2b
|
musashi RNA-binding protein 2b |
| chr21_+_22423286 | 0.19 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr12_+_15002757 | 0.19 |
ENSDART00000135036
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr9_+_41080029 | 0.19 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr7_+_21887787 | 0.18 |
ENSDART00000162252
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr17_+_6276559 | 0.18 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr25_-_32311048 | 0.18 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr3_+_49397115 | 0.18 |
ENSDART00000176042
|
tecra
|
trans-2,3-enoyl-CoA reductase a |
| chr25_+_34889061 | 0.18 |
ENSDART00000136226
|
spire2
|
spire-type actin nucleation factor 2 |
| chr20_-_36679233 | 0.18 |
ENSDART00000062908
|
rpl7l1
|
ribosomal protein L7-like 1 |
| chr14_-_677801 | 0.18 |
ENSDART00000158505
|
LO018568.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.2 | 1.0 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 0.5 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.8 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.4 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 0.4 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.4 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.5 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.5 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 0.4 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.4 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.2 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.9 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.1 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.0 | 0.1 | GO:0034138 | chemokine production(GO:0032602) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 1.0 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.4 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0006747 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.0 | 0.4 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) folic acid metabolic process(GO:0046655) |
| 0.0 | 0.2 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.0 | 0.3 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.2 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 0.0 | 0.1 | GO:0034397 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:2001057 | reactive nitrogen species metabolic process(GO:2001057) |
| 0.0 | 0.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.2 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.0 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 1.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 1.0 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.1 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 0.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.8 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.9 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.1 | GO:0035545 | determination of left/right asymmetry in diencephalon(GO:0035462) determination of left/right asymmetry in nervous system(GO:0035545) |
| 0.0 | 0.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.1 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.2 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.1 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.6 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.0 | 0.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 1.0 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.2 | 0.8 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 1.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 1.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.5 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.4 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.4 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 1.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.5 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.2 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.1 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.3 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.0 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 1.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.0 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |