Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
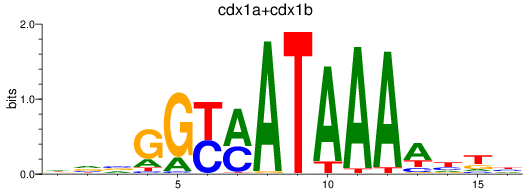
Results for cdx1a+cdx1b
Z-value: 1.00
Transcription factors associated with cdx1a+cdx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cdx1a
|
ENSDARG00000114554 | caudal type homeobox 1a |
|
cdx1b
|
ENSDARG00000116139 | caudal type homeobox 1 b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cdx1b | dr11_v1_chr21_-_43992027_43992027 | -0.88 | 5.1e-07 | Click! |
| cdx1a | dr11_v1_chr14_+_3287740_3287740 | -0.75 | 2.1e-04 | Click! |
Activity profile of cdx1a+cdx1b motif
Sorted Z-values of cdx1a+cdx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_25257022 | 2.22 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr20_-_29499363 | 2.18 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr19_-_47571456 | 2.16 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr3_-_15999501 | 2.13 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr7_-_17028015 | 2.10 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr12_-_13205854 | 1.94 |
ENSDART00000077829
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr19_+_1872794 | 1.92 |
ENSDART00000013217
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr8_-_32803227 | 1.90 |
ENSDART00000110079
|
zgc:194839
|
zgc:194839 |
| chr11_-_25257045 | 1.89 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr25_-_29074064 | 1.85 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr6_+_40629066 | 1.80 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr11_-_25257595 | 1.77 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr19_+_1873059 | 1.74 |
ENSDART00000145246
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr9_-_48370645 | 1.70 |
ENSDART00000140185
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr10_-_10864331 | 1.67 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr1_-_53468160 | 1.64 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr16_+_38940758 | 1.62 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr21_-_39024754 | 1.59 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr23_-_43718067 | 1.57 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr17_-_51222510 | 1.56 |
ENSDART00000166687
|
psen1
|
presenilin 1 |
| chr2_-_9744081 | 1.56 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr10_-_31563049 | 1.50 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr2_+_19777146 | 1.49 |
ENSDART00000038648
|
ptbp2b
|
polypyrimidine tract binding protein 2b |
| chr23_+_31596441 | 1.44 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr19_-_10306910 | 1.44 |
ENSDART00000151684
|
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr12_+_13205955 | 1.42 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr13_+_11436130 | 1.40 |
ENSDART00000169895
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr15_+_834697 | 1.35 |
ENSDART00000154861
ENSDART00000160694 |
si:dkey-7i4.14
zgc:174573
|
si:dkey-7i4.14 zgc:174573 |
| chr5_-_40510397 | 1.34 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr3_+_60589157 | 1.34 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
| chr17_+_43595692 | 1.33 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr23_+_44349252 | 1.33 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr21_-_11970199 | 1.31 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr17_+_32343121 | 1.30 |
ENSDART00000156051
|
dhx32b
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32b |
| chr1_-_14506759 | 1.30 |
ENSDART00000057044
|
si:dkey-194g4.1
|
si:dkey-194g4.1 |
| chr11_+_6116503 | 1.29 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr2_-_6115389 | 1.28 |
ENSDART00000134921
|
prdx1
|
peroxiredoxin 1 |
| chr23_+_36087219 | 1.27 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr14_+_14841685 | 1.27 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr9_+_32178374 | 1.26 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr1_+_41596099 | 1.25 |
ENSDART00000111367
|
si:dkey-56e3.3
|
si:dkey-56e3.3 |
| chr24_-_3407507 | 1.25 |
ENSDART00000132648
|
nck1b
|
NCK adaptor protein 1b |
| chr5_+_38662125 | 1.23 |
ENSDART00000136949
|
si:dkey-58f10.13
|
si:dkey-58f10.13 |
| chr23_-_3758637 | 1.22 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr21_-_25416391 | 1.22 |
ENSDART00000114081
|
sms
|
spermine synthase |
| chr24_+_39105051 | 1.22 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr19_+_27970292 | 1.21 |
ENSDART00000103887
ENSDART00000145549 |
med10
|
mediator complex subunit 10 |
| chr8_-_39822917 | 1.20 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr5_-_69499486 | 1.19 |
ENSDART00000023983
ENSDART00000180293 |
psat1
|
phosphoserine aminotransferase 1 |
| chr21_+_19070921 | 1.16 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr19_+_19747430 | 1.16 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr9_+_22782027 | 1.14 |
ENSDART00000090816
|
rif1
|
replication timing regulatory factor 1 |
| chr7_-_59589547 | 1.12 |
ENSDART00000008554
ENSDART00000101731 |
larp7
|
La ribonucleoprotein domain family, member 7 |
| chr16_+_52966812 | 1.12 |
ENSDART00000148435
ENSDART00000049099 ENSDART00000150117 |
trip13
|
thyroid hormone receptor interactor 13 |
| chr5_-_36597612 | 1.12 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr10_-_31562695 | 1.12 |
ENSDART00000186456
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr7_+_50766094 | 1.11 |
ENSDART00000165037
|
si:ch73-380l10.2
|
si:ch73-380l10.2 |
| chr6_-_7107868 | 1.09 |
ENSDART00000171123
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr22_+_10440991 | 1.08 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr18_+_9171778 | 1.08 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr23_+_7379728 | 1.08 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr10_-_3332362 | 1.07 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr8_-_50888806 | 1.06 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr14_+_35369979 | 1.03 |
ENSDART00000144702
ENSDART00000169712 |
clint1a
|
clathrin interactor 1a |
| chr10_-_21545091 | 1.02 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr14_-_45604667 | 1.02 |
ENSDART00000167016
|
si:ch211-276i12.9
|
si:ch211-276i12.9 |
| chr14_+_21754521 | 1.02 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr7_-_48263516 | 1.01 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr24_-_24114217 | 1.01 |
ENSDART00000160578
|
zgc:112982
|
zgc:112982 |
| chr9_+_2041535 | 1.01 |
ENSDART00000093187
|
lnpa
|
limb and neural patterns a |
| chr21_+_41468856 | 1.00 |
ENSDART00000161175
|
bod1
|
biorientation of chromosomes in cell division 1 |
| chr16_+_29376751 | 1.00 |
ENSDART00000168856
ENSDART00000162502 ENSDART00000050535 |
rrnad1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr6_-_23002373 | 0.99 |
ENSDART00000037709
ENSDART00000170369 |
nol11
|
nucleolar protein 11 |
| chr14_+_4276394 | 0.99 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr3_+_62161184 | 0.99 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr8_-_25033681 | 0.98 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr21_-_15929041 | 0.97 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr1_+_45663727 | 0.96 |
ENSDART00000038574
ENSDART00000141144 ENSDART00000149565 |
trappc5
|
trafficking protein particle complex 5 |
| chr16_-_17586883 | 0.95 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr23_+_43718115 | 0.94 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr4_-_56898328 | 0.94 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr6_-_6976096 | 0.94 |
ENSDART00000151822
ENSDART00000039443 ENSDART00000177960 |
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr10_-_44924289 | 0.92 |
ENSDART00000171267
|
tuba7l
|
tubulin, alpha 7 like |
| chr1_+_27808916 | 0.92 |
ENSDART00000102335
|
pspc1
|
paraspeckle component 1 |
| chr5_+_70155935 | 0.92 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr12_+_27022517 | 0.92 |
ENSDART00000152975
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr5_+_64856666 | 0.92 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr14_+_34490445 | 0.91 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr12_-_33746111 | 0.90 |
ENSDART00000127203
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr23_-_24699400 | 0.89 |
ENSDART00000104031
|
sdf4
|
stromal cell derived factor 4 |
| chr16_+_42471455 | 0.89 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr22_+_2512154 | 0.89 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr22_-_9930079 | 0.88 |
ENSDART00000131734
|
si:dkey-253d23.9
|
si:dkey-253d23.9 |
| chr21_+_41468210 | 0.86 |
ENSDART00000192287
|
bod1
|
biorientation of chromosomes in cell division 1 |
| chr2_+_23351454 | 0.85 |
ENSDART00000017034
ENSDART00000154645 |
rnf2
|
ring finger protein 2 |
| chr3_+_22442445 | 0.85 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr20_-_34670236 | 0.84 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr2_-_6115688 | 0.83 |
ENSDART00000081663
|
prdx1
|
peroxiredoxin 1 |
| chr10_+_15340768 | 0.82 |
ENSDART00000046274
ENSDART00000168909 |
trappc13
|
trafficking protein particle complex 13 |
| chr3_-_4501026 | 0.82 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr14_-_2602445 | 0.81 |
ENSDART00000166910
|
etf1a
|
eukaryotic translation termination factor 1a |
| chr4_-_5775507 | 0.80 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr5_-_31856681 | 0.79 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr5_-_1963498 | 0.79 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr10_+_27068251 | 0.79 |
ENSDART00000012717
|
ehd1b
|
EH-domain containing 1b |
| chr2_-_32366287 | 0.78 |
ENSDART00000144758
|
ubtfl
|
upstream binding transcription factor, like |
| chr15_+_30323491 | 0.77 |
ENSDART00000048847
|
nos2b
|
nitric oxide synthase 2b, inducible |
| chr19_-_17304866 | 0.77 |
ENSDART00000160433
ENSDART00000190798 ENSDART00000171284 |
sf3a3
|
splicing factor 3a, subunit 3 |
| chr3_+_36646054 | 0.75 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr6_-_10728921 | 0.75 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr20_-_43741159 | 0.75 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr2_+_19785540 | 0.74 |
ENSDART00000149789
|
ptbp2b
|
polypyrimidine tract binding protein 2b |
| chr18_-_12295092 | 0.74 |
ENSDART00000033248
|
fam107b
|
family with sequence similarity 107, member B |
| chr19_+_19652439 | 0.73 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr2_-_27975530 | 0.73 |
ENSDART00000020367
|
zgc:56556
|
zgc:56556 |
| chr6_-_37745508 | 0.71 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr1_+_46194333 | 0.71 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr23_+_36115541 | 0.70 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr24_+_5237753 | 0.70 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr10_-_34889053 | 0.69 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr12_+_3871452 | 0.68 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr9_-_16853462 | 0.68 |
ENSDART00000160273
|
CT573248.2
|
|
| chr4_-_20135919 | 0.68 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr1_+_47178529 | 0.67 |
ENSDART00000158432
ENSDART00000074450 ENSDART00000137448 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr2_+_23039041 | 0.67 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr1_-_45752010 | 0.67 |
ENSDART00000101410
|
si:ch211-214c7.5
|
si:ch211-214c7.5 |
| chr4_-_42014474 | 0.66 |
ENSDART00000193544
|
si:dkey-237m9.1
|
si:dkey-237m9.1 |
| chr23_-_10722664 | 0.66 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr22_+_2403068 | 0.66 |
ENSDART00000132925
ENSDART00000132569 |
zgc:112977
|
zgc:112977 |
| chr24_-_17029374 | 0.65 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr3_+_24190207 | 0.65 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr3_+_39579393 | 0.64 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr10_+_40321067 | 0.63 |
ENSDART00000109816
|
gltpb
|
glycolipid transfer protein b |
| chr5_-_31857345 | 0.62 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr13_+_45431660 | 0.62 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr22_+_21255860 | 0.62 |
ENSDART00000134893
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr2_+_26682913 | 0.62 |
ENSDART00000010683
ENSDART00000131411 ENSDART00000137933 |
impa1
|
inositol monophosphatase 1 |
| chr22_-_25043103 | 0.61 |
ENSDART00000015512
|
polr3f
|
polymerase (RNA) III (DNA directed) polypeptide F |
| chr8_+_25959940 | 0.60 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr7_+_18989241 | 0.59 |
ENSDART00000053213
|
zgc:194252
|
zgc:194252 |
| chr24_-_10014512 | 0.59 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr19_+_3840955 | 0.59 |
ENSDART00000172305
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr22_-_10605045 | 0.58 |
ENSDART00000184812
|
bap1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr18_+_14684115 | 0.57 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr16_-_21915223 | 0.57 |
ENSDART00000170634
|
setdb1b
|
SET domain, bifurcated 1b |
| chr9_+_51891 | 0.57 |
ENSDART00000163529
|
zgc:158316
|
zgc:158316 |
| chr11_-_25461336 | 0.56 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr16_-_9675982 | 0.56 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr16_-_25368048 | 0.56 |
ENSDART00000132445
|
rbfa
|
ribosome binding factor A |
| chr11_-_18323059 | 0.56 |
ENSDART00000182590
|
SFMBT1
|
Scm like with four mbt domains 1 |
| chr23_-_39959173 | 0.56 |
ENSDART00000077122
|
xcr1a.1
|
chemokine (C motif) receptor 1a, duplicate 1 |
| chr11_-_36051004 | 0.55 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr11_-_44622478 | 0.54 |
ENSDART00000171837
|
tbce
|
tubulin folding cofactor E |
| chr17_-_30652738 | 0.54 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr18_-_14691727 | 0.54 |
ENSDART00000010129
|
pdf
|
peptide deformylase, mitochondrial |
| chr16_-_16225260 | 0.54 |
ENSDART00000165790
|
gra
|
granulito |
| chr10_+_40629616 | 0.54 |
ENSDART00000147476
|
CR396590.9
|
|
| chr24_-_10006158 | 0.53 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr10_+_26612321 | 0.53 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr2_+_35733335 | 0.52 |
ENSDART00000113489
|
rasal2
|
RAS protein activator like 2 |
| chr5_+_50913034 | 0.52 |
ENSDART00000149787
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr14_+_15495088 | 0.51 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr7_+_52154215 | 0.50 |
ENSDART00000098712
|
TMEM208
|
zgc:77041 |
| chr12_+_3262564 | 0.48 |
ENSDART00000184264
|
tmem101
|
transmembrane protein 101 |
| chr23_-_39959784 | 0.48 |
ENSDART00000115046
|
xcr1a.1
|
chemokine (C motif) receptor 1a, duplicate 1 |
| chr3_-_53091946 | 0.48 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr21_+_25236297 | 0.48 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr5_+_58687541 | 0.47 |
ENSDART00000083015
ENSDART00000181902 |
ccdc84
|
coiled-coil domain containing 84 |
| chr11_+_44622472 | 0.47 |
ENSDART00000159068
ENSDART00000166323 ENSDART00000187753 |
rbm34
|
RNA binding motif protein 34 |
| chr15_+_42285643 | 0.46 |
ENSDART00000152731
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr2_+_16798923 | 0.46 |
ENSDART00000181603
ENSDART00000179816 ENSDART00000087107 ENSDART00000187009 |
eif4g1a
|
eukaryotic translation initiation factor 4 gamma, 1a |
| chr1_+_8508753 | 0.45 |
ENSDART00000091683
|
alkbh5
|
alkB homolog 5, RNA demethylase |
| chr22_-_10440688 | 0.44 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr17_-_43666166 | 0.44 |
ENSDART00000077990
|
egr2a
|
early growth response 2a |
| chr25_+_9013342 | 0.44 |
ENSDART00000154207
ENSDART00000153705 |
im:7145024
|
im:7145024 |
| chr20_-_13140309 | 0.43 |
ENSDART00000020703
ENSDART00000188594 |
ints7
|
integrator complex subunit 7 |
| chr21_+_20396858 | 0.42 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr23_+_309099 | 0.42 |
ENSDART00000134556
ENSDART00000055152 |
taf11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr10_+_8481660 | 0.41 |
ENSDART00000109559
|
BX682234.1
|
|
| chr2_-_37458527 | 0.40 |
ENSDART00000146820
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr24_-_9997948 | 0.40 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr20_+_27087539 | 0.40 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr14_-_15171435 | 0.40 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr13_+_18520738 | 0.40 |
ENSDART00000113952
|
tlr4al
|
toll-like receptor 4a, like |
| chr1_-_7603734 | 0.40 |
ENSDART00000009315
|
mxb
|
myxovirus (influenza) resistance B |
| chr1_-_9486214 | 0.39 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr21_-_30181732 | 0.39 |
ENSDART00000015636
|
hnrnph1l
|
heterogeneous nuclear ribonucleoprotein H1, like |
| chr5_-_12713920 | 0.38 |
ENSDART00000099749
|
CLDN22
|
si:dkey-98f17.3 |
| chr16_+_34046733 | 0.38 |
ENSDART00000058424
|
fam46ba
|
family with sequence similarity 46, member Ba |
| chr4_+_25630555 | 0.38 |
ENSDART00000133425
|
acot15
|
acyl-CoA thioesterase 15 |
| chr12_-_31726748 | 0.37 |
ENSDART00000153174
|
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr16_-_27138478 | 0.36 |
ENSDART00000147438
|
tmem245
|
transmembrane protein 245 |
| chr2_+_19522082 | 0.36 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr6_+_49551614 | 0.36 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr15_+_9297340 | 0.36 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr3_-_4557653 | 0.36 |
ENSDART00000111305
|
ftr50
|
finTRIM family, member 50 |
| chr2_+_31948352 | 0.36 |
ENSDART00000192611
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr16_+_43344475 | 0.36 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr14_+_22447662 | 0.35 |
ENSDART00000161776
|
sowahab
|
sosondowah ankyrin repeat domain family member Ab |
| chr5_-_24997293 | 0.35 |
ENSDART00000177478
|
emid1
|
EMI domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cdx1a+cdx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.4 | 1.2 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.4 | 1.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.4 | 1.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.4 | 4.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 1.6 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.4 | 1.1 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.4 | 2.1 | GO:0097065 | anterior head development(GO:0097065) |
| 0.3 | 1.0 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.3 | 1.0 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.3 | 1.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 1.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.3 | 1.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.3 | 1.0 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.2 | 2.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 1.0 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 1.9 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 1.6 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 1.1 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.2 | 2.1 | GO:0098869 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 2.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 0.9 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 1.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 0.9 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.2 | 1.1 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 3.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 0.5 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.2 | 0.8 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.7 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 2.1 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.8 | GO:0031284 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.7 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 3.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.5 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.6 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 1.9 | GO:0033753 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 0.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.0 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.1 | 0.6 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.2 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.1 | 2.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 1.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:0060958 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 1.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.9 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 0.2 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.5 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 1.3 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.0 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.6 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 2.1 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.9 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.8 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 1.1 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 0.9 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.4 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.1 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 3.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.8 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 1.3 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.3 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 2.8 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 2.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.5 | 3.7 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.3 | 1.6 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.3 | 1.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 1.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 0.9 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 1.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 1.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.2 | 0.5 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.2 | 1.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 1.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.6 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 2.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 3.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.1 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.0 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 2.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.7 | GO:0005938 | cell cortex(GO:0005938) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.6 | 4.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 1.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 1.0 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.3 | 2.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 0.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 0.6 | GO:0052833 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 1.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 0.7 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 1.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 1.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.6 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.5 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 1.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.8 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 1.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 2.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 1.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 2.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 1.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.9 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 2.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.1 | GO:0043566 | structure-specific DNA binding(GO:0043566) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.9 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 3.2 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.0 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 1.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 1.6 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 3.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |