Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
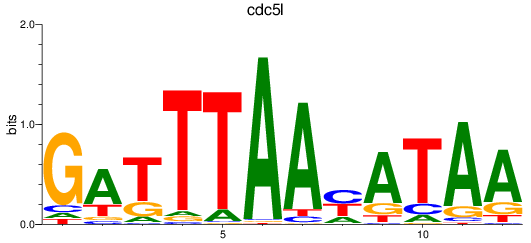
Results for cdc5l
Z-value: 2.46
Transcription factors associated with cdc5l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cdc5l
|
ENSDARG00000043797 | CDC5 cell division cycle 5-like (S. pombe) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cdc5l | dr11_v1_chr17_+_5061135_5061135 | 0.82 | 1.6e-05 | Click! |
Activity profile of cdc5l motif
Sorted Z-values of cdc5l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_29509133 | 7.20 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr5_-_61652254 | 6.83 |
ENSDART00000097364
|
drl
|
draculin |
| chr5_+_50879545 | 5.90 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr21_-_44512893 | 5.90 |
ENSDART00000166853
|
zgc:136410
|
zgc:136410 |
| chr11_+_41981959 | 5.55 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr7_+_24881680 | 5.49 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr20_-_26491567 | 5.42 |
ENSDART00000147154
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr8_-_16725573 | 5.36 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr9_+_34397516 | 5.14 |
ENSDART00000011304
ENSDART00000192973 |
med14
|
mediator complex subunit 14 |
| chr4_+_35553514 | 4.96 |
ENSDART00000182938
|
CR847906.1
|
|
| chr20_-_38801981 | 4.66 |
ENSDART00000125333
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr7_-_17337233 | 4.61 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr5_-_61624693 | 4.47 |
ENSDART00000141323
|
si:dkey-261j4.4
|
si:dkey-261j4.4 |
| chr2_-_42958619 | 4.47 |
ENSDART00000144317
|
oc90
|
otoconin 90 |
| chr3_+_6443992 | 4.36 |
ENSDART00000169325
ENSDART00000162255 |
nup85
|
nucleoporin 85 |
| chr17_+_5985933 | 4.13 |
ENSDART00000190844
|
zgc:194275
|
zgc:194275 |
| chr19_+_27859546 | 4.10 |
ENSDART00000161908
|
nsun2
|
NOP2/Sun RNA methyltransferase family, member 2 |
| chr11_-_6880725 | 3.87 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr2_-_42960353 | 3.84 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr16_-_25699554 | 3.83 |
ENSDART00000155899
ENSDART00000114495 |
ddx61
|
DEAD (Asp-Glu-Ala-Asp) box helicase 61 |
| chr1_-_11104805 | 3.79 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr6_-_8377055 | 3.78 |
ENSDART00000131513
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr22_-_16291041 | 3.74 |
ENSDART00000021666
|
rtca
|
RNA 3'-terminal phosphate cyclase |
| chr9_-_27398369 | 3.72 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr2_-_19234329 | 3.67 |
ENSDART00000161106
ENSDART00000160060 ENSDART00000174552 |
cdc20
|
cell division cycle 20 homolog |
| chr15_-_16333015 | 3.67 |
ENSDART00000080892
|
txndc17
|
thioredoxin domain containing 17 |
| chr9_+_34397843 | 3.64 |
ENSDART00000146314
|
med14
|
mediator complex subunit 14 |
| chr19_-_13933204 | 3.64 |
ENSDART00000158059
|
trnau1apa
|
tRNA selenocysteine 1 associated protein 1a |
| chr6_+_60125033 | 3.61 |
ENSDART00000148557
ENSDART00000008224 |
aurka
|
aurora kinase A |
| chr23_-_23179417 | 3.53 |
ENSDART00000122945
ENSDART00000091662 |
noc2l
|
NOC2-like nucleolar associated transcriptional repressor |
| chr23_+_32011768 | 3.52 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr7_-_20464133 | 3.46 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr17_-_31695217 | 3.31 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr4_-_11580948 | 3.31 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr3_+_23697997 | 3.29 |
ENSDART00000184299
ENSDART00000078466 |
hoxb3a
|
homeobox B3a |
| chr17_-_14700889 | 3.28 |
ENSDART00000179975
|
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr21_+_20901505 | 3.23 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr4_-_71110826 | 3.23 |
ENSDART00000167431
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr14_-_30905963 | 3.22 |
ENSDART00000183543
ENSDART00000186441 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr24_+_22485710 | 3.21 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr13_-_37474989 | 3.21 |
ENSDART00000114136
|
wdr89
|
WD repeat domain 89 |
| chr2_+_2168547 | 3.19 |
ENSDART00000029347
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr9_-_21976670 | 3.17 |
ENSDART00000104322
|
uchl3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr9_+_44994214 | 3.16 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr13_+_18321140 | 3.16 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr11_-_33868881 | 3.12 |
ENSDART00000163295
ENSDART00000172633 ENSDART00000171439 |
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr21_-_22357545 | 3.11 |
ENSDART00000134320
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr1_-_38171648 | 3.09 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr6_+_29386728 | 3.08 |
ENSDART00000104303
|
actl6a
|
actin-like 6A |
| chr16_-_12060770 | 3.07 |
ENSDART00000183237
ENSDART00000103948 |
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr8_+_45003659 | 3.06 |
ENSDART00000132663
|
si:ch211-163b2.4
|
si:ch211-163b2.4 |
| chr7_-_24994568 | 3.06 |
ENSDART00000002961
|
rcor2
|
REST corepressor 2 |
| chr12_+_1609563 | 3.04 |
ENSDART00000163559
|
SLC39A11
|
solute carrier family 39 member 11 |
| chr4_+_17642731 | 3.04 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr21_-_29990574 | 3.03 |
ENSDART00000185800
|
si:ch73-190f9.4
|
si:ch73-190f9.4 |
| chr15_-_14038227 | 3.02 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr24_-_3426620 | 3.00 |
ENSDART00000184346
|
nck1b
|
NCK adaptor protein 1b |
| chr5_+_45007962 | 2.99 |
ENSDART00000010786
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr4_-_56068511 | 2.99 |
ENSDART00000168345
|
znf1133
|
zinc finger protein 1133 |
| chr25_+_19557628 | 2.96 |
ENSDART00000133859
|
lyrm5b
|
LYR motif containing 5b |
| chr2_-_23390779 | 2.96 |
ENSDART00000020136
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr16_+_55059026 | 2.91 |
ENSDART00000109391
|
LO017815.1
|
Danio rerio nuclear receptor coactivator 7-like (LOC792958), mRNA. |
| chr21_-_29533723 | 2.91 |
ENSDART00000193554
|
BX537120.2
|
|
| chr19_+_16064439 | 2.89 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr22_+_8313513 | 2.88 |
ENSDART00000181169
ENSDART00000103911 |
CABZ01077217.1
|
|
| chr4_+_18441988 | 2.87 |
ENSDART00000040827
|
ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr19_-_3056235 | 2.87 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr16_-_12060488 | 2.85 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr17_-_24937879 | 2.84 |
ENSDART00000153964
|
CR391986.1
|
|
| chr2_-_2813259 | 2.84 |
ENSDART00000032540
|
usp14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr18_-_18874921 | 2.84 |
ENSDART00000193332
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr22_+_1911269 | 2.82 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr24_-_9294134 | 2.82 |
ENSDART00000082434
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr13_+_33606739 | 2.82 |
ENSDART00000026464
|
cfl1l
|
cofilin 1 (non-muscle), like |
| chr25_-_3217115 | 2.79 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr14_+_22132388 | 2.79 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr7_-_20464468 | 2.79 |
ENSDART00000134700
|
cnpy4
|
canopy4 |
| chr7_-_24994722 | 2.78 |
ENSDART00000131671
|
rcor2
|
REST corepressor 2 |
| chr20_+_28803642 | 2.78 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr20_+_43691208 | 2.77 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr10_+_22918338 | 2.76 |
ENSDART00000167874
ENSDART00000171298 |
zgc:103508
|
zgc:103508 |
| chr17_-_8312923 | 2.75 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr20_-_27225876 | 2.75 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr1_-_53880639 | 2.72 |
ENSDART00000010543
|
ltv1
|
LTV1 ribosome biogenesis factor |
| chr24_-_31904924 | 2.71 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr19_+_636886 | 2.69 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr9_-_32300783 | 2.66 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr21_-_29735366 | 2.65 |
ENSDART00000181668
|
CR847571.2
|
|
| chr9_+_30090656 | 2.65 |
ENSDART00000102981
|
col8a1a
|
collagen, type VIII, alpha 1a |
| chr8_+_22355909 | 2.64 |
ENSDART00000146457
ENSDART00000142883 |
zgc:153631
|
zgc:153631 |
| chr21_-_29689141 | 2.64 |
ENSDART00000100876
|
CR847571.1
|
|
| chr19_+_15444210 | 2.63 |
ENSDART00000142509
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr21_-_19919020 | 2.63 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr5_+_33289057 | 2.61 |
ENSDART00000123210
|
med22
|
mediator complex subunit 22 |
| chr6_-_30683637 | 2.59 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr19_-_8536474 | 2.57 |
ENSDART00000139715
|
dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr16_-_33650578 | 2.57 |
ENSDART00000058460
|
utp11l
|
UTP11-like, U3 small nucleolar ribonucleoprotein (yeast) |
| chr4_+_23127284 | 2.54 |
ENSDART00000122675
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr6_-_19340889 | 2.52 |
ENSDART00000181407
|
mif4gda
|
MIF4G domain containing a |
| chr6_-_29402578 | 2.51 |
ENSDART00000115031
|
mrpl47
|
mitochondrial ribosomal protein L47 |
| chr10_+_13209580 | 2.51 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr14_+_36220479 | 2.50 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr4_+_64562090 | 2.50 |
ENSDART00000188810
|
si:ch211-223a21.3
|
si:ch211-223a21.3 |
| chr9_-_41040098 | 2.49 |
ENSDART00000008275
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr5_+_45008418 | 2.47 |
ENSDART00000189882
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr2_+_50967947 | 2.46 |
ENSDART00000162288
|
si:ch211-249o11.5
|
si:ch211-249o11.5 |
| chr7_-_26844064 | 2.46 |
ENSDART00000162241
|
si:ch211-107p11.3
|
si:ch211-107p11.3 |
| chr3_-_54607166 | 2.46 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr13_+_30912117 | 2.44 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr6_-_41091151 | 2.43 |
ENSDART00000154963
ENSDART00000153818 |
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr18_-_18875308 | 2.41 |
ENSDART00000127182
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr24_-_38657683 | 2.41 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr9_-_32300611 | 2.41 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr7_-_28647959 | 2.39 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr18_+_6536598 | 2.38 |
ENSDART00000149350
|
fkbp4
|
FK506 binding protein 4 |
| chr2_-_10877228 | 2.38 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr21_-_29477624 | 2.35 |
ENSDART00000180110
|
BX537120.1
|
|
| chr3_-_42125655 | 2.34 |
ENSDART00000040753
|
nudt1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr20_+_26394324 | 2.32 |
ENSDART00000078093
|
zbtb2b
|
zinc finger and BTB domain containing 2b |
| chr2_-_9607879 | 2.30 |
ENSDART00000056899
|
txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr12_-_13336703 | 2.30 |
ENSDART00000134356
|
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_-_30762264 | 2.28 |
ENSDART00000085454
|
dis3
|
DIS3 exosome endoribonuclease and 3'-5' exoribonuclease |
| chr6_-_52234796 | 2.26 |
ENSDART00000001803
|
tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr18_+_6536293 | 2.25 |
ENSDART00000024576
|
fkbp4
|
FK506 binding protein 4 |
| chr13_-_42757565 | 2.24 |
ENSDART00000161950
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr10_+_16036246 | 2.24 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr5_+_60847823 | 2.24 |
ENSDART00000074426
|
lig3
|
ligase III, DNA, ATP-dependent |
| chr24_+_22022109 | 2.24 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr16_+_29690708 | 2.23 |
ENSDART00000103054
|
lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr23_-_29553430 | 2.22 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr9_+_21259820 | 2.22 |
ENSDART00000137024
ENSDART00000132324 |
ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr11_-_36350421 | 2.22 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr8_-_16697912 | 2.21 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr21_-_29940453 | 2.21 |
ENSDART00000182862
|
CU104716.1
|
|
| chr17_-_36529449 | 2.20 |
ENSDART00000187252
|
colec11
|
collectin sub-family member 11 |
| chr19_+_20163826 | 2.20 |
ENSDART00000090942
ENSDART00000134650 |
ccdc126
|
coiled-coil domain containing 126 |
| chr19_+_3206263 | 2.19 |
ENSDART00000020344
|
zgc:86598
|
zgc:86598 |
| chr25_+_5249513 | 2.19 |
ENSDART00000126814
|
CABZ01039863.1
|
|
| chr4_-_29769304 | 2.19 |
ENSDART00000134990
|
znf1119
|
zinc finger protein 1119 |
| chr3_-_24681404 | 2.18 |
ENSDART00000161612
|
BX569789.1
|
|
| chr6_-_19341184 | 2.18 |
ENSDART00000168236
ENSDART00000167674 |
mif4gda
|
MIF4G domain containing a |
| chr15_-_41689981 | 2.17 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr23_-_3409140 | 2.17 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr18_+_17537975 | 2.17 |
ENSDART00000179783
|
nup93
|
nucleoporin 93 |
| chr4_-_76488854 | 2.16 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr5_-_41645058 | 2.16 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr10_+_16036573 | 2.16 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr19_-_27588842 | 2.15 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr4_+_69863750 | 2.15 |
ENSDART00000170800
|
znf1117
|
zinc finger protein 1117 |
| chr2_-_57264262 | 2.15 |
ENSDART00000183815
ENSDART00000149829 ENSDART00000088508 ENSDART00000149508 |
mbd3a
|
methyl-CpG binding domain protein 3a |
| chr4_+_29532731 | 2.13 |
ENSDART00000114649
|
znf1140
|
zinc finger protein 1140 |
| chr10_+_39893439 | 2.12 |
ENSDART00000003435
|
smfn
|
small fragment nuclease |
| chr22_-_15593824 | 2.12 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr6_-_37745508 | 2.11 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr12_-_9533956 | 2.10 |
ENSDART00000188940
|
nudt13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr10_+_21434649 | 2.10 |
ENSDART00000193938
ENSDART00000064558 |
etf1b
|
eukaryotic translation termination factor 1b |
| chr14_-_38826739 | 2.07 |
ENSDART00000187633
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr18_-_48296793 | 2.07 |
ENSDART00000032184
ENSDART00000193076 |
CABZ01069595.1
|
|
| chr19_-_27578929 | 2.07 |
ENSDART00000177368
|
si:dkeyp-46h3.3
|
si:dkeyp-46h3.3 |
| chr11_-_16152400 | 2.06 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr25_-_15040369 | 2.06 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr4_+_1761029 | 2.05 |
ENSDART00000026963
|
pmpcb
|
peptidase (mitochondrial processing) beta |
| chr12_+_6065661 | 2.05 |
ENSDART00000142418
|
sgms1
|
sphingomyelin synthase 1 |
| chr15_-_20024205 | 2.04 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr2_-_32666174 | 2.04 |
ENSDART00000133660
|
puf60a
|
poly-U binding splicing factor a |
| chr20_-_25645150 | 2.04 |
ENSDART00000063137
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr3_+_62161184 | 2.03 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr11_+_24994705 | 2.03 |
ENSDART00000129211
|
zgc:92107
|
zgc:92107 |
| chr3_-_61375496 | 2.02 |
ENSDART00000165188
|
si:dkey-111k8.2
|
si:dkey-111k8.2 |
| chr18_-_17087138 | 2.01 |
ENSDART00000135597
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr14_+_22132896 | 1.99 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr11_+_16152316 | 1.99 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr23_-_29553691 | 1.99 |
ENSDART00000053804
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr7_+_39664055 | 1.99 |
ENSDART00000146171
|
zgc:158564
|
zgc:158564 |
| chr2_-_22530969 | 1.98 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr3_+_32651697 | 1.98 |
ENSDART00000055338
|
thoc6
|
THO complex 6 |
| chr2_+_52049239 | 1.97 |
ENSDART00000036813
|
ccdc94
|
coiled-coil domain containing 94 |
| chr1_+_34695373 | 1.97 |
ENSDART00000077744
ENSDART00000102118 ENSDART00000135528 |
gtf2f2a
|
general transcription factor IIF, polypeptide 2a |
| chr8_-_28274251 | 1.96 |
ENSDART00000050671
|
rap1aa
|
RAP1A, member of RAS oncogene family a |
| chr6_+_43426599 | 1.96 |
ENSDART00000056457
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr4_+_30454496 | 1.94 |
ENSDART00000164555
|
si:dkey-199m13.4
|
si:dkey-199m13.4 |
| chr9_+_29430432 | 1.94 |
ENSDART00000125632
|
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr11_+_1533097 | 1.93 |
ENSDART00000066191
|
ift52
|
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr11_-_16395956 | 1.92 |
ENSDART00000115085
|
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr17_-_19463355 | 1.92 |
ENSDART00000045881
|
dicer1
|
dicer 1, ribonuclease type III |
| chr4_-_77260727 | 1.91 |
ENSDART00000075770
|
zgc:162948
|
zgc:162948 |
| chr17_-_36529016 | 1.91 |
ENSDART00000025019
|
colec11
|
collectin sub-family member 11 |
| chr25_+_17925424 | 1.90 |
ENSDART00000067305
|
zgc:103499
|
zgc:103499 |
| chr6_+_3716666 | 1.90 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr12_+_5708400 | 1.89 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr18_+_14633974 | 1.89 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr11_+_24815667 | 1.88 |
ENSDART00000141730
|
rabif
|
RAB interacting factor |
| chr7_-_8315179 | 1.88 |
ENSDART00000184049
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr4_+_33225161 | 1.87 |
ENSDART00000150851
|
si:dkey-247i3.5
|
si:dkey-247i3.5 |
| chr23_-_13295923 | 1.87 |
ENSDART00000189214
ENSDART00000165432 |
CR450832.1
|
Danio rerio uncharacterized LOC100001444 (LOC100001444), mRNA. |
| chr13_+_22555342 | 1.87 |
ENSDART00000193633
|
bmpr1aa
|
bone morphogenetic protein receptor, type IAa |
| chr5_+_18047111 | 1.87 |
ENSDART00000132164
|
hira
|
histone cell cycle regulator a |
| chr17_-_15189397 | 1.85 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr2_+_2169337 | 1.85 |
ENSDART00000179939
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr4_+_45504471 | 1.84 |
ENSDART00000150399
|
si:dkey-256i11.2
|
si:dkey-256i11.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cdc5l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 1.6 | 4.7 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 1.5 | 4.6 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 1.3 | 5.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 1.2 | 3.6 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.9 | 1.9 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.9 | 2.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.9 | 2.7 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.9 | 2.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.8 | 5.0 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.7 | 2.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.7 | 2.2 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.7 | 2.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.7 | 2.8 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.7 | 1.4 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.7 | 9.1 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.6 | 1.9 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.6 | 1.9 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.6 | 3.7 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.6 | 5.9 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.6 | 1.7 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.6 | 2.2 | GO:0016117 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.5 | 1.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.5 | 3.3 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.5 | 1.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.5 | 3.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.5 | 1.6 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.5 | 2.5 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.5 | 6.6 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 0.5 | 1.5 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.5 | 2.5 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.5 | 2.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.5 | 2.9 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.5 | 3.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.5 | 1.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.5 | 2.3 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.5 | 1.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.4 | 1.3 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.4 | 1.3 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.4 | 2.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.4 | 1.7 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.4 | 2.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 0.8 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.4 | 1.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 1.6 | GO:0071962 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.4 | 1.6 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.4 | 2.0 | GO:0071320 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 1.1 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 4.1 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.4 | 1.1 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.4 | 5.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.4 | 1.4 | GO:0042148 | strand invasion(GO:0042148) |
| 0.4 | 3.5 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.4 | 2.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 1.4 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.3 | 1.0 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.3 | 2.3 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.3 | 1.0 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.3 | 1.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.3 | 0.9 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.3 | 2.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 1.8 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.3 | 2.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.3 | 8.8 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.3 | 2.0 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.3 | 0.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.3 | 2.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.3 | 0.8 | GO:0046133 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.3 | 2.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 | 1.8 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.3 | 1.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.2 | 2.0 | GO:0036372 | opsin transport(GO:0036372) |
| 0.2 | 2.7 | GO:0014028 | notochord formation(GO:0014028) |
| 0.2 | 0.7 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.2 | 0.9 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.2 | 0.2 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.2 | 2.3 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.2 | 1.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 0.7 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 2.6 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.2 | 3.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 1.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 1.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 2.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.2 | 2.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 5.1 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.2 | 2.1 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.2 | 0.8 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.2 | 1.7 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 1.9 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 1.7 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.2 | 2.8 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 1.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 3.3 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.2 | 1.8 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 1.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.2 | 0.4 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.2 | 0.7 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.2 | 2.8 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.2 | 1.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 1.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 6.2 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.2 | 4.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.2 | 5.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 0.5 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.2 | 5.2 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.2 | 1.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.2 | 5.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 3.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 2.9 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.1 | 1.8 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.8 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 1.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.7 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.6 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.2 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.1 | 1.5 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 2.8 | GO:0003128 | heart field specification(GO:0003128) |
| 0.1 | 0.7 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 7.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 0.6 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 2.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.7 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.1 | 1.0 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 4.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.3 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 1.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.9 | GO:0051279 | regulation of calcium ion transport into cytosol(GO:0010522) regulation of release of sequestered calcium ion into cytosol(GO:0051279) |
| 0.1 | 0.8 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 1.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 2.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.5 | GO:0045161 | neuronal ion channel clustering(GO:0045161) clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 1.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.7 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.1 | 2.6 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.1 | 1.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 2.7 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 1.6 | GO:0060324 | face development(GO:0060324) |
| 0.1 | 3.1 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.8 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.1 | 1.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 2.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 1.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 7.9 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 0.9 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 1.8 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.5 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.1 | 0.4 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 2.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 7.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 6.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.1 | 1.8 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.5 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 1.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 5.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 1.5 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.1 | 0.2 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 1.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.0 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.9 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 2.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 2.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.2 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 | 3.1 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.1 | 0.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 3.0 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.7 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 4.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 8.4 | GO:0016072 | rRNA processing(GO:0006364) rRNA metabolic process(GO:0016072) |
| 0.1 | 0.6 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.8 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 3.4 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.8 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) regulation of phospholipid metabolic process(GO:1903725) |
| 0.1 | 7.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.9 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 2.0 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 2.7 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.1 | 0.6 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 0.4 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.8 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 2.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 2.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 4.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.3 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 1.1 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 4.4 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.8 | GO:0042744 | oxygen transport(GO:0015671) hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 3.0 | GO:0006417 | regulation of translation(GO:0006417) |
| 0.0 | 1.1 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) positive regulation of protein modification by small protein conjugation or removal(GO:1903322) |
| 0.0 | 1.6 | GO:0002218 | activation of innate immune response(GO:0002218) pattern recognition receptor signaling pathway(GO:0002221) toll-like receptor signaling pathway(GO:0002224) innate immune response-activating signal transduction(GO:0002758) |
| 0.0 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 1.3 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.3 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.2 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 1.0 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.9 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.9 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 1.0 | GO:2000027 | regulation of organ morphogenesis(GO:2000027) |
| 0.0 | 1.0 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 1.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 1.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.6 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.6 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 2.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.5 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.6 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 1.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) positive regulation of cellular protein catabolic process(GO:1903364) |
| 0.0 | 4.2 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 1.1 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.7 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 1.6 | GO:0048565 | digestive tract development(GO:0048565) |
| 0.0 | 1.3 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.8 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 3.6 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 4.4 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.5 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 1.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.4 | GO:0009913 | epidermal cell differentiation(GO:0009913) |
| 0.0 | 0.8 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.8 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 2.2 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 5.7 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.0 | 3.8 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 1.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.7 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 2.7 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.3 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.8 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.7 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.5 | GO:0045446 | endothelial cell differentiation(GO:0045446) |
| 0.0 | 0.2 | GO:0006000 | fructose metabolic process(GO:0006000) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 1.3 | 8.8 | GO:0070847 | core mediator complex(GO:0070847) |
| 1.2 | 3.5 | GO:0030689 | Noc complex(GO:0030689) |
| 1.0 | 2.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.9 | 2.7 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.9 | 2.6 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.7 | 2.8 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.6 | 1.9 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.6 | 3.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.6 | 2.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.5 | 5.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.5 | 1.6 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.5 | 2.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.4 | 1.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.4 | 2.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 4.6 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.4 | 1.6 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.4 | 4.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 2.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.4 | 1.9 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.4 | 2.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 1.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 2.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 1.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 1.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.3 | 4.9 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.3 | 4.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 2.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.3 | 2.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.3 | 1.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.3 | 10.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 4.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 3.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 2.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 4.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 1.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 2.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 1.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.2 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 0.6 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.2 | 4.1 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 0.7 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.2 | 1.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 1.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 4.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 7.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 2.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 1.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 0.5 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.2 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.2 | 0.5 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.1 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 7.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 4.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.5 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 0.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.5 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 1.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 2.2 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.7 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.3 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.1 | 0.9 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 1.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.7 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 3.1 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 4.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 1.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.1 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 3.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 0.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 3.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 5.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.5 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 4.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 7.5 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 3.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.8 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 1.0 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 9.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 5.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 3.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 3.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 1.3 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 1.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 100.6 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 1.5 | 6.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.4 | 4.1 | GO:0042806 | fucose binding(GO:0042806) |
| 1.2 | 6.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 1.2 | 4.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 1.0 | 4.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.9 | 2.7 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.8 | 2.5 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.8 | 5.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.7 | 3.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.7 | 2.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.7 | 2.8 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.7 | 2.0 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.6 | 4.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.6 | 2.9 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.6 | 1.7 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.6 | 1.7 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.6 | 2.2 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.5 | 4.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 1.6 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.5 | 3.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.5 | 1.5 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.5 | 1.5 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.5 | 1.9 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.5 | 1.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.5 | 1.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 3.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.4 | 1.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.4 | 1.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.4 | 2.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 2.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.4 | 9.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.3 | 2.0 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 1.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.3 | 1.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 4.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.3 | 0.9 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 1.2 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.3 | 1.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 3.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 1.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.3 | 7.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 1.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 0.7 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 2.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 2.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 2.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 2.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 1.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 1.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 2.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 2.3 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.2 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 2.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 0.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 0.7 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.5 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.2 | 1.4 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 1.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.5 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.2 | 2.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.2 | 1.6 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 5.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 3.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.4 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.2 | 2.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.7 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.7 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 1.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 5.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 2.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.0 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.6 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.6 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 0.6 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 3.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.8 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 9.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 4.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 3.3 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.1 | 2.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 2.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 3.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.8 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 3.3 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 1.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 2.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 1.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 8.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 2.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 2.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 10.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 3.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 7.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.4 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.8 | GO:0008144 | drug binding(GO:0008144) |
| 0.1 | 3.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 9.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 1.2 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 2.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 2.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 1.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 3.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 74.3 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 1.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 2.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.6 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.7 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 6.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.2 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 3.2 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 14.1 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 13.2 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 4.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 3.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 1.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.2 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 16.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 12.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 9.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 6.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 7.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 4.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 1.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 5.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 3.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 1.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.3 | 1.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 2.5 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.3 | 4.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 2.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 1.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 4.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.2 | 1.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 4.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 1.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 7.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 10.4 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 2.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 6.8 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.2 | 6.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 3.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 6.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 13.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 1.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 1.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 2.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.2 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 6.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 5.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.7 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.1 | 3.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 3.8 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 6.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.0 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 1.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 3.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |