Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
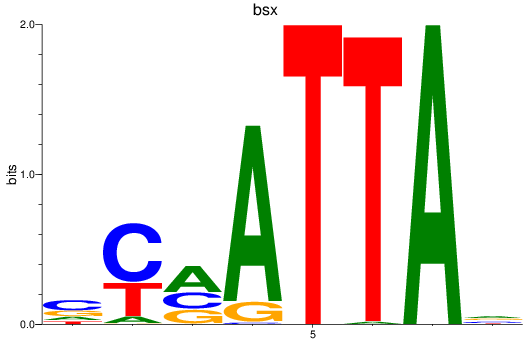
Results for bsx
Z-value: 0.50
Transcription factors associated with bsx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bsx
|
ENSDARG00000068976 | brain-specific homeobox |
|
bsx
|
ENSDARG00000110782 | brain-specific homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bsx | dr11_v1_chr10_-_29892486_29892486 | 0.41 | 8.0e-02 | Click! |
Activity profile of bsx motif
Sorted Z-values of bsx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_50888806 | 0.92 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr8_+_30452945 | 0.80 |
ENSDART00000062303
|
foxd5
|
forkhead box D5 |
| chr16_+_29509133 | 0.75 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr18_-_33979693 | 0.74 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr19_+_15440841 | 0.74 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr23_+_38251864 | 0.73 |
ENSDART00000183498
ENSDART00000129593 |
znf217
|
zinc finger protein 217 |
| chr22_+_18315490 | 0.73 |
ENSDART00000160413
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr21_-_37973819 | 0.71 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr24_+_39638555 | 0.69 |
ENSDART00000078313
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr15_-_20233105 | 0.68 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr5_+_50879545 | 0.68 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr17_-_31659670 | 0.63 |
ENSDART00000030448
|
vsx2
|
visual system homeobox 2 |
| chr25_+_14507567 | 0.61 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr15_+_1796313 | 0.59 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr7_-_54679595 | 0.58 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr19_+_15441022 | 0.58 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_+_20793982 | 0.56 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr1_+_23563691 | 0.56 |
ENSDART00000142879
|
ncapg
|
non-SMC condensin I complex, subunit G |
| chr17_+_21295132 | 0.56 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr5_+_68807170 | 0.55 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr10_-_35257458 | 0.55 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr6_+_41191482 | 0.54 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr2_-_26596794 | 0.51 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr6_-_43283122 | 0.51 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr2_-_20599315 | 0.49 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr12_-_42214 | 0.48 |
ENSDART00000045071
|
foxk2
|
forkhead box K2 |
| chr3_-_26183699 | 0.47 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr1_-_55248496 | 0.47 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr11_+_18130300 | 0.46 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr25_+_18965430 | 0.46 |
ENSDART00000169742
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr22_-_14272699 | 0.46 |
ENSDART00000190121
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr12_-_5728755 | 0.45 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr3_+_18807006 | 0.45 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr3_+_26244353 | 0.44 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr23_-_18913032 | 0.43 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr10_-_44560165 | 0.43 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr15_-_16177603 | 0.43 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr17_-_28101880 | 0.42 |
ENSDART00000149458
ENSDART00000149226 ENSDART00000085758 |
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr13_-_42400647 | 0.42 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr3_-_16719244 | 0.42 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr8_+_21353878 | 0.42 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr3_-_26184018 | 0.42 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr3_+_23737795 | 0.42 |
ENSDART00000182247
|
hoxb3a
|
homeobox B3a |
| chr16_+_28994709 | 0.42 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr2_+_4146606 | 0.42 |
ENSDART00000171170
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr18_+_17537344 | 0.41 |
ENSDART00000025782
|
nup93
|
nucleoporin 93 |
| chr1_-_33645967 | 0.41 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr25_-_29074064 | 0.40 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr8_-_4892913 | 0.40 |
ENSDART00000126808
|
igsf9b
|
immunoglobulin superfamily, member 9b |
| chr12_-_26415499 | 0.39 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr10_+_22775253 | 0.39 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr2_+_4146299 | 0.39 |
ENSDART00000173418
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr21_-_37194365 | 0.39 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr24_+_24170914 | 0.38 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr9_-_24031461 | 0.38 |
ENSDART00000021218
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr5_+_66433287 | 0.37 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr2_-_55298075 | 0.37 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr9_-_27748868 | 0.37 |
ENSDART00000190306
|
tbccd1
|
TBCC domain containing 1 |
| chr3_+_6469754 | 0.37 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr19_-_7540821 | 0.37 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr12_-_35830625 | 0.37 |
ENSDART00000180028
|
CU459056.1
|
|
| chr9_-_20372977 | 0.37 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr19_-_19871211 | 0.36 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr12_+_16087077 | 0.36 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr4_+_9177997 | 0.36 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr15_+_1534644 | 0.36 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr15_-_14884332 | 0.35 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr12_-_26430507 | 0.35 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr5_+_1624359 | 0.35 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr17_+_16046132 | 0.35 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr18_+_29898955 | 0.35 |
ENSDART00000064080
|
cenpn
|
centromere protein N |
| chr23_-_40194732 | 0.35 |
ENSDART00000164931
|
tgm1l2
|
transglutaminase 1 like 2 |
| chr23_-_31913231 | 0.35 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr4_+_13428993 | 0.34 |
ENSDART00000067151
|
si:dkey-39a18.1
|
si:dkey-39a18.1 |
| chr23_-_31913069 | 0.34 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr17_+_16046314 | 0.34 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr13_-_11984867 | 0.34 |
ENSDART00000157538
|
npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr2_+_33326522 | 0.34 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr21_-_17296789 | 0.34 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr1_-_12064715 | 0.34 |
ENSDART00000143628
ENSDART00000103406 |
pla2g12a
|
phospholipase A2, group XIIA |
| chr11_+_18157260 | 0.33 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr8_+_48484455 | 0.33 |
ENSDART00000122737
|
PRDM16
|
si:ch211-263k4.2 |
| chr13_-_21660203 | 0.33 |
ENSDART00000100925
|
mxtx1
|
mix-type homeobox gene 1 |
| chr3_-_21288202 | 0.33 |
ENSDART00000191766
ENSDART00000187319 |
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr7_+_56098590 | 0.33 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr8_-_20243389 | 0.32 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr11_+_18175893 | 0.32 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr6_-_40922971 | 0.31 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr12_+_31638045 | 0.31 |
ENSDART00000184216
ENSDART00000183645 ENSDART00000153129 |
dnmbp
|
dynamin binding protein |
| chr21_-_43666420 | 0.31 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr11_-_6974022 | 0.31 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr5_-_41307550 | 0.31 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr4_+_14981854 | 0.31 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr9_-_21460164 | 0.31 |
ENSDART00000133469
|
zmym2
|
zinc finger, MYM-type 2 |
| chr21_+_43404945 | 0.30 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr2_-_32666174 | 0.30 |
ENSDART00000133660
|
puf60a
|
poly-U binding splicing factor a |
| chr23_+_11285662 | 0.30 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr18_+_17537975 | 0.30 |
ENSDART00000179783
|
nup93
|
nucleoporin 93 |
| chr12_-_33817114 | 0.30 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr19_+_15521997 | 0.30 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr19_+_19786117 | 0.30 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr2_+_56891858 | 0.30 |
ENSDART00000159912
|
zgc:85843
|
zgc:85843 |
| chr17_-_10043273 | 0.29 |
ENSDART00000156078
|
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr17_-_20118145 | 0.29 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr18_-_14777092 | 0.29 |
ENSDART00000144660
|
mtss1la
|
metastasis suppressor 1-like a |
| chr21_+_34088110 | 0.29 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr23_+_36095260 | 0.29 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr13_-_12005429 | 0.29 |
ENSDART00000180302
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr13_+_17468161 | 0.29 |
ENSDART00000008906
|
znf503
|
zinc finger protein 503 |
| chr5_+_27897504 | 0.28 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr2_-_38206034 | 0.28 |
ENSDART00000144518
ENSDART00000137395 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr1_+_41588170 | 0.28 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr11_-_36341028 | 0.28 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr16_+_42471455 | 0.27 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr20_-_23426339 | 0.27 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr3_+_13848226 | 0.27 |
ENSDART00000184342
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr10_-_41980797 | 0.27 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr23_-_1017605 | 0.27 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr15_+_36309070 | 0.27 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr5_+_60590796 | 0.27 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr24_-_10021341 | 0.26 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr3_+_60721342 | 0.26 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr23_-_25135046 | 0.26 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr17_-_49412313 | 0.26 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr25_-_27722614 | 0.26 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr24_+_37800102 | 0.26 |
ENSDART00000187591
|
telo2
|
TEL2, telomere maintenance 2, homolog (S. cerevisiae) |
| chr21_-_13149453 | 0.26 |
ENSDART00000172578
|
si:dkey-228b2.6
|
si:dkey-228b2.6 |
| chr19_+_31585917 | 0.26 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr9_+_32073606 | 0.26 |
ENSDART00000184170
ENSDART00000180355 ENSDART00000110204 ENSDART00000123278 |
pikfyve
|
phosphoinositide kinase, FYVE finger containing |
| chr5_-_41142467 | 0.26 |
ENSDART00000129415
|
zfr
|
zinc finger RNA binding protein |
| chr25_+_35553542 | 0.26 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr5_+_38913621 | 0.26 |
ENSDART00000137112
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr1_+_33668236 | 0.25 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr5_-_68333081 | 0.25 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr7_+_17816006 | 0.25 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr21_-_36396334 | 0.25 |
ENSDART00000183627
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr21_+_28478663 | 0.25 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr10_-_23099809 | 0.25 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr3_+_28860283 | 0.25 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr8_-_39822917 | 0.25 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr5_-_31856681 | 0.24 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr17_-_25649079 | 0.24 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr22_-_27709692 | 0.24 |
ENSDART00000172458
|
CR547131.1
|
|
| chr11_+_24703108 | 0.24 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr7_+_17816470 | 0.24 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr24_-_4450238 | 0.24 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr10_-_28380919 | 0.24 |
ENSDART00000183409
ENSDART00000183105 ENSDART00000100207 ENSDART00000185392 ENSDART00000131220 |
btg3
|
B-cell translocation gene 3 |
| chr1_+_34696503 | 0.24 |
ENSDART00000186106
|
CR339054.2
|
|
| chr5_-_65691512 | 0.24 |
ENSDART00000189255
|
notch1b
|
notch 1b |
| chr21_-_30994577 | 0.24 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr4_+_40265583 | 0.23 |
ENSDART00000151870
|
si:ch211-246n15.2
|
si:ch211-246n15.2 |
| chr7_+_53498152 | 0.23 |
ENSDART00000184497
|
znf609b
|
zinc finger protein 609b |
| chr17_-_31695217 | 0.23 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr19_+_16015881 | 0.23 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr5_+_30998764 | 0.23 |
ENSDART00000185783
ENSDART00000147874 |
ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr20_-_48898560 | 0.23 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr18_+_9171778 | 0.23 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr12_-_8504278 | 0.23 |
ENSDART00000135865
|
egr2b
|
early growth response 2b |
| chr14_+_24845941 | 0.23 |
ENSDART00000187513
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr19_-_20403507 | 0.23 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr13_+_30903816 | 0.23 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr14_-_21959712 | 0.23 |
ENSDART00000021417
|
p2rx3a
|
purinergic receptor P2X, ligand-gated ion channel, 3a |
| chr11_-_29563437 | 0.23 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr2_+_41526904 | 0.23 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr6_+_50393047 | 0.22 |
ENSDART00000055502
ENSDART00000055511 |
ergic3
|
ERGIC and golgi 3 |
| chr19_+_12406583 | 0.22 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr13_+_27232848 | 0.22 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr8_-_23612462 | 0.22 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr13_+_36984794 | 0.22 |
ENSDART00000137328
|
frmd6
|
FERM domain containing 6 |
| chr10_+_26944418 | 0.22 |
ENSDART00000135493
|
frmd8
|
FERM domain containing 8 |
| chr17_+_24821627 | 0.21 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr18_-_25568994 | 0.21 |
ENSDART00000133029
|
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr4_-_74892355 | 0.21 |
ENSDART00000188725
|
PHF21B
|
PHD finger protein 21B |
| chr20_+_46513651 | 0.21 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr16_+_16266428 | 0.21 |
ENSDART00000188433
|
setd2
|
SET domain containing 2 |
| chr21_+_33187992 | 0.21 |
ENSDART00000162745
ENSDART00000188388 |
BX072577.1
|
|
| chr15_-_23206562 | 0.21 |
ENSDART00000110360
|
ccdc153
|
coiled-coil domain containing 153 |
| chr6_+_15762647 | 0.21 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr10_+_17371356 | 0.21 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr9_+_50316921 | 0.21 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr18_+_24921587 | 0.21 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr3_+_23738215 | 0.21 |
ENSDART00000143981
|
hoxb3a
|
homeobox B3a |
| chr7_+_17908235 | 0.21 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr7_-_17816175 | 0.21 |
ENSDART00000091272
ENSDART00000173757 |
ecsit
|
ECSIT signalling integrator |
| chr10_+_518546 | 0.21 |
ENSDART00000128275
|
npffr1l3
|
neuropeptide FF receptor 1 like 3 |
| chr7_-_51368681 | 0.21 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr7_+_29863548 | 0.21 |
ENSDART00000136555
ENSDART00000134068 |
tln2a
|
talin 2a |
| chr8_-_16788626 | 0.20 |
ENSDART00000191652
|
CR759968.2
|
|
| chr21_+_30746348 | 0.20 |
ENSDART00000050172
|
trpc2b
|
transient receptor potential cation channel, subfamily C, member 2b |
| chr15_+_436776 | 0.20 |
ENSDART00000008504
|
med17
|
mediator complex subunit 17 |
| chr16_+_14707960 | 0.20 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr16_-_41714988 | 0.20 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr11_-_18557929 | 0.20 |
ENSDART00000110882
ENSDART00000181381 ENSDART00000189312 |
dido1
|
death inducer-obliterator 1 |
| chr10_+_23099890 | 0.20 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr5_-_12063381 | 0.20 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr3_+_62205858 | 0.20 |
ENSDART00000126807
|
zgc:173575
|
zgc:173575 |
| chr4_-_2545310 | 0.20 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr3_-_26244256 | 0.19 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr23_+_39695827 | 0.19 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr2_-_42071558 | 0.19 |
ENSDART00000142792
|
cspp1b
|
centrosome and spindle pole associated protein 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of bsx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.2 | 0.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 0.6 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.2 | 0.8 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 0.5 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.4 | GO:2000726 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.4 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.5 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.6 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.4 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.4 | GO:0051661 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.3 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.2 | GO:0060898 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.2 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.4 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.6 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 0.4 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.2 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.3 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.3 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.9 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.7 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.6 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.5 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.2 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 1.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.2 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0030910 | olfactory placode formation(GO:0030910) |
| 0.0 | 0.3 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.3 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.3 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.3 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0060584 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 0.1 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:1900158 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.0 | 0.2 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.6 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.5 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.3 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.3 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 1.2 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.9 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.6 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.9 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.3 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.5 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.2 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.7 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.4 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0098847 | double-stranded telomeric DNA binding(GO:0003691) single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.0 | GO:0031005 | filamin binding(GO:0031005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |