Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
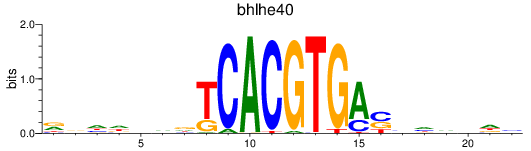
Results for bhlhe40
Z-value: 0.65
Transcription factors associated with bhlhe40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bhlhe40
|
ENSDARG00000004060 | basic helix-loop-helix family, member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bhlhe40 | dr11_v1_chr11_-_35763323_35763323 | 0.77 | 1.0e-04 | Click! |
Activity profile of bhlhe40 motif
Sorted Z-values of bhlhe40 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_54765262 | 1.55 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr7_+_48999723 | 1.52 |
ENSDART00000182699
ENSDART00000166329 |
si:ch211-288d18.1
|
si:ch211-288d18.1 |
| chr25_-_35599887 | 1.40 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr4_+_13696537 | 1.12 |
ENSDART00000109195
ENSDART00000122041 ENSDART00000192554 |
nrcama
|
neuronal cell adhesion molecule a |
| chr10_-_44027391 | 1.01 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr10_-_6775271 | 0.79 |
ENSDART00000110735
|
zgc:194281
|
zgc:194281 |
| chr16_+_26824691 | 0.70 |
ENSDART00000135053
|
zmp:0000001316
|
zmp:0000001316 |
| chr21_+_43178831 | 0.63 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr24_+_7631797 | 0.62 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr19_+_40379771 | 0.61 |
ENSDART00000017917
ENSDART00000110699 |
vps50
vps50
|
VPS50 EARP/GARPII complex subunit VPS50 EARP/GARPII complex subunit |
| chr20_+_40766645 | 0.58 |
ENSDART00000144401
|
tbc1d32
|
TBC1 domain family, member 32 |
| chr23_+_42131509 | 0.57 |
ENSDART00000184544
|
SHISAL2A
|
shisa like 2A |
| chr3_-_25420931 | 0.57 |
ENSDART00000109601
ENSDART00000182184 |
bptf
|
bromodomain PHD finger transcription factor |
| chr6_-_49547680 | 0.56 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr9_+_36946340 | 0.55 |
ENSDART00000135281
|
si:dkey-3d4.3
|
si:dkey-3d4.3 |
| chr15_-_44077937 | 0.54 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr17_+_10501647 | 0.52 |
ENSDART00000140391
|
tyro3
|
TYRO3 protein tyrosine kinase |
| chr13_+_19884631 | 0.51 |
ENSDART00000089533
|
atrnl1a
|
attractin-like 1a |
| chr24_-_42072886 | 0.49 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr9_-_1200187 | 0.49 |
ENSDART00000158760
|
ino80da
|
INO80 complex subunit Da |
| chr15_+_59417 | 0.49 |
ENSDART00000187260
|
AXL
|
AXL receptor tyrosine kinase |
| chr10_-_35108683 | 0.48 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr15_-_41290415 | 0.47 |
ENSDART00000152157
|
si:dkey-75b17.1
|
si:dkey-75b17.1 |
| chr4_+_331351 | 0.46 |
ENSDART00000132625
|
tulp4a
|
tubby like protein 4a |
| chr20_+_46213553 | 0.45 |
ENSDART00000100532
|
stx7l
|
syntaxin 7-like |
| chr21_+_4540127 | 0.44 |
ENSDART00000043431
|
nup188
|
nucleoporin 188 |
| chr20_+_13969414 | 0.43 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr12_-_3778848 | 0.41 |
ENSDART00000152128
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr3_-_39627412 | 0.39 |
ENSDART00000123292
|
si:dkey-27o4.1
|
si:dkey-27o4.1 |
| chr21_-_4539899 | 0.39 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr8_+_17933475 | 0.39 |
ENSDART00000100651
|
erich3
|
glutamate-rich 3 |
| chr7_-_67214972 | 0.38 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr12_-_7280551 | 0.38 |
ENSDART00000061633
|
zgc:171971
|
zgc:171971 |
| chr22_-_16759151 | 0.36 |
ENSDART00000191880
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr7_+_1442059 | 0.35 |
ENSDART00000173391
|
si:cabz01090193.1
|
si:cabz01090193.1 |
| chr15_-_41807371 | 0.33 |
ENSDART00000156819
|
slc25a36b
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36b |
| chr14_-_25452503 | 0.33 |
ENSDART00000148652
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr20_-_45040916 | 0.33 |
ENSDART00000190001
|
klhl29
|
kelch-like family member 29 |
| chr3_-_25421504 | 0.33 |
ENSDART00000154200
|
bptf
|
bromodomain PHD finger transcription factor |
| chr17_-_37299394 | 0.32 |
ENSDART00000154414
|
ptgr2
|
prostaglandin reductase 2 |
| chr12_+_47044707 | 0.31 |
ENSDART00000186506
|
zranb1a
|
zinc finger, RAN-binding domain containing 1a |
| chr12_+_13652361 | 0.31 |
ENSDART00000182757
ENSDART00000152689 |
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr24_-_21498802 | 0.31 |
ENSDART00000181235
ENSDART00000153695 |
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr1_+_50639416 | 0.31 |
ENSDART00000141977
|
herc3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr18_+_6479963 | 0.30 |
ENSDART00000092752
ENSDART00000136333 |
wash1
|
WAS protein family homolog 1 |
| chr22_+_30184039 | 0.29 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr10_+_26515946 | 0.28 |
ENSDART00000134276
|
synj1
|
synaptojanin 1 |
| chr15_-_34418525 | 0.28 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr5_+_32009956 | 0.28 |
ENSDART00000188482
|
scai
|
suppressor of cancer cell invasion |
| chr22_-_16758973 | 0.27 |
ENSDART00000145208
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr15_+_1534644 | 0.27 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr12_+_13652747 | 0.27 |
ENSDART00000066359
|
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr18_+_39074139 | 0.26 |
ENSDART00000142390
|
gnb5b
|
guanine nucleotide binding protein (G protein), beta 5b |
| chr3_+_13929860 | 0.25 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr15_-_26570948 | 0.25 |
ENSDART00000156621
|
wdr81
|
WD repeat domain 81 |
| chr12_+_17933775 | 0.24 |
ENSDART00000186047
ENSDART00000160586 |
trrap
|
transformation/transcription domain-associated protein |
| chr19_-_5103313 | 0.23 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr19_+_791538 | 0.23 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr9_-_10805231 | 0.23 |
ENSDART00000193913
ENSDART00000078348 |
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr4_+_17655872 | 0.23 |
ENSDART00000066999
|
washc3
|
WASH complex subunit 3 |
| chr7_-_14446512 | 0.22 |
ENSDART00000041577
|
kif7
|
kinesin family member 7 |
| chr16_+_25296389 | 0.22 |
ENSDART00000114528
|
tbc1d31
|
TBC1 domain family, member 31 |
| chr10_-_15644904 | 0.22 |
ENSDART00000138389
ENSDART00000101191 ENSDART00000186559 ENSDART00000122170 |
smc5
|
structural maintenance of chromosomes 5 |
| chr12_+_45677293 | 0.22 |
ENSDART00000152850
ENSDART00000153047 |
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr17_-_44440832 | 0.22 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr24_+_3307857 | 0.21 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr9_-_34269066 | 0.21 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr16_+_5251768 | 0.20 |
ENSDART00000144558
|
plecb
|
plectin b |
| chr5_-_66798729 | 0.18 |
ENSDART00000113077
|
FERMT3 (1 of many)
|
im:7154036 |
| chr12_-_44010532 | 0.18 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr15_+_1148074 | 0.18 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr18_-_7677208 | 0.16 |
ENSDART00000092456
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr14_+_94946 | 0.16 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr7_+_65939098 | 0.16 |
ENSDART00000193599
|
RASSF10
|
Ras association domain family member 10 |
| chr19_-_38830582 | 0.16 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr8_-_6943155 | 0.15 |
ENSDART00000139545
ENSDART00000033294 |
wdr13
|
WD repeat domain 13 |
| chr23_-_2901167 | 0.15 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr15_+_34939906 | 0.15 |
ENSDART00000131182
|
si:ch73-95l15.3
|
si:ch73-95l15.3 |
| chr10_-_24724388 | 0.15 |
ENSDART00000148582
|
smpd1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr11_-_44163164 | 0.15 |
ENSDART00000047126
|
clcn4
|
chloride channel, voltage-sensitive 4 |
| chr17_+_32622933 | 0.14 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr23_-_31913231 | 0.13 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr16_+_19029297 | 0.12 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr21_+_45816030 | 0.12 |
ENSDART00000187056
|
pitx1
|
paired-like homeodomain 1 |
| chr15_+_6652396 | 0.12 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr25_-_1323623 | 0.11 |
ENSDART00000156532
ENSDART00000157163 ENSDART00000156062 ENSDART00000082447 ENSDART00000189175 |
calml4b
|
calmodulin-like 4b |
| chr9_+_54686686 | 0.11 |
ENSDART00000066198
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr20_-_51100669 | 0.11 |
ENSDART00000023488
|
atp6v1d
|
ATPase H+ transporting V1 subunit D |
| chr14_-_3071564 | 0.10 |
ENSDART00000166136
ENSDART00000167138 |
slc35a4
|
solute carrier family 35, member A4 |
| chr20_-_211920 | 0.09 |
ENSDART00000104790
|
znf292b
|
zinc finger protein 292b |
| chr4_-_5831522 | 0.08 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr3_+_12784460 | 0.08 |
ENSDART00000168382
|
cyp2k8
|
cytochrome P450, family 2, subfamily K, polypeptide 8 |
| chr1_+_54766943 | 0.07 |
ENSDART00000144759
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr21_-_14251306 | 0.07 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr5_+_41476443 | 0.06 |
ENSDART00000145228
ENSDART00000137981 ENSDART00000142538 |
pias2
|
protein inhibitor of activated STAT, 2 |
| chr11_-_236984 | 0.06 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr21_+_11916788 | 0.05 |
ENSDART00000136103
|
ubap2a
|
ubiquitin associated protein 2a |
| chr22_+_8800432 | 0.05 |
ENSDART00000139940
|
si:dkey-182g1.6
|
si:dkey-182g1.6 |
| chr23_+_9035578 | 0.05 |
ENSDART00000140455
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr7_+_34620418 | 0.05 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr1_-_38538284 | 0.04 |
ENSDART00000009777
|
glra3
|
glycine receptor, alpha 3 |
| chr23_+_32028574 | 0.03 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr19_+_9111550 | 0.03 |
ENSDART00000088336
|
setdb1a
|
SET domain, bifurcated 1a |
| chr13_+_40692804 | 0.02 |
ENSDART00000109822
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr5_-_9540641 | 0.02 |
ENSDART00000124384
ENSDART00000160079 |
gak
|
cyclin G associated kinase |
| chr14_-_45595711 | 0.02 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr18_+_31280984 | 0.01 |
ENSDART00000170285
ENSDART00000150608 ENSDART00000159720 |
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr6_+_12968101 | 0.01 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr5_+_26204561 | 0.01 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr13_+_12389849 | 0.01 |
ENSDART00000086525
|
atp10d
|
ATPase phospholipid transporting 10D |
Network of associatons between targets according to the STRING database.
First level regulatory network of bhlhe40
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.4 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 0.3 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.6 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.3 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.2 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.6 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.3 | GO:0032474 | sulfate transport(GO:0008272) otolith morphogenesis(GO:0032474) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 1.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0035082 | axoneme assembly(GO:0035082) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.2 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |