Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
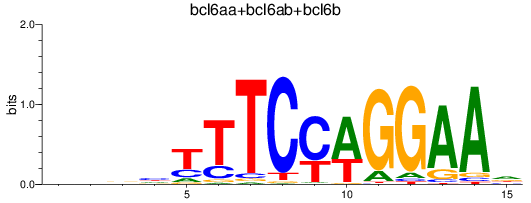
Results for bcl6aa+bcl6ab+bcl6b
Z-value: 1.15
Transcription factors associated with bcl6aa+bcl6ab+bcl6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bcl6ab
|
ENSDARG00000069295 | BCL6A transcription repressor b |
|
bcl6b
|
ENSDARG00000069335 | BCL6B transcription repressor |
|
bcl6aa
|
ENSDARG00000070864 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000111395 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000112502 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000116260 | BCL6A transcription repressor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bcl6b | dr11_v1_chr7_+_20019125_20019125 | -0.92 | 1.8e-08 | Click! |
| bcl6ab | dr11_v1_chr2_-_10098191_10098191 | -0.78 | 9.5e-05 | Click! |
| bcl6a | dr11_v1_chr6_-_28222592_28222592 | -0.38 | 1.0e-01 | Click! |
Activity profile of bcl6aa+bcl6ab+bcl6b motif
Sorted Z-values of bcl6aa+bcl6ab+bcl6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_52061034 | 4.15 |
ENSDART00000170383
|
CABZ01089777.1
|
|
| chr16_-_52879741 | 3.20 |
ENSDART00000166470
|
TPPP
|
tubulin polymerization promoting protein |
| chr16_-_36834505 | 2.85 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr11_-_8167799 | 2.61 |
ENSDART00000133574
ENSDART00000024046 ENSDART00000146940 |
uox
|
urate oxidase |
| chr17_-_6730247 | 2.33 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr6_-_345503 | 2.29 |
ENSDART00000168901
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr20_+_10166297 | 2.27 |
ENSDART00000141877
|
kcnk10a
|
potassium channel, subfamily K, member 10a |
| chr24_-_40009446 | 2.06 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr6_+_9881093 | 2.01 |
ENSDART00000151558
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr25_-_17395315 | 2.01 |
ENSDART00000064596
|
cyp2x8
|
cytochrome P450, family 2, subfamily X, polypeptide 8 |
| chr8_-_23783633 | 2.00 |
ENSDART00000132657
|
si:ch211-163l21.7
|
si:ch211-163l21.7 |
| chr24_+_42948 | 1.92 |
ENSDART00000122785
|
tmx3b
|
thioredoxin related transmembrane protein 3b |
| chr23_-_35483163 | 1.89 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr22_+_7480465 | 1.76 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr5_-_3209345 | 1.75 |
ENSDART00000171477
|
CABZ01076737.1
|
|
| chr17_-_24564674 | 1.74 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr7_-_12869545 | 1.53 |
ENSDART00000163045
|
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr3_+_49021079 | 1.53 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr18_+_49411417 | 1.51 |
ENSDART00000028944
|
LRFN3
|
zmp:0000001073 |
| chr24_+_35947077 | 1.51 |
ENSDART00000173406
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr1_+_44053641 | 1.50 |
ENSDART00000124873
|
si:ch73-109d9.4
|
si:ch73-109d9.4 |
| chr6_-_35446110 | 1.50 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr21_+_11684830 | 1.40 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr9_-_21912227 | 1.40 |
ENSDART00000145576
|
lmo7a
|
LIM domain 7a |
| chr21_-_18982337 | 1.37 |
ENSDART00000147246
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr15_+_16521785 | 1.36 |
ENSDART00000062191
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr21_+_11685009 | 1.36 |
ENSDART00000014668
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr16_+_45746549 | 1.36 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr9_-_6372535 | 1.34 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr15_-_40246396 | 1.34 |
ENSDART00000063777
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr23_+_45229198 | 1.33 |
ENSDART00000172445
|
ttc39b
|
tetratricopeptide repeat domain 39B |
| chr11_-_762721 | 1.32 |
ENSDART00000166465
|
syn2b
|
synapsin IIb |
| chr5_+_63668735 | 1.27 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr4_-_18436899 | 1.25 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr11_-_22303678 | 1.24 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr3_+_12861158 | 1.23 |
ENSDART00000171237
|
kcnj2b
|
potassium inwardly-rectifying channel, subfamily J, member 2b |
| chr7_+_38509333 | 1.21 |
ENSDART00000153482
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr25_-_20258508 | 1.20 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr6_+_9175886 | 1.19 |
ENSDART00000165333
|
si:ch211-207l14.1
|
si:ch211-207l14.1 |
| chr18_+_50907675 | 1.18 |
ENSDART00000159950
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr23_+_29966466 | 1.16 |
ENSDART00000143583
|
dvl1a
|
dishevelled segment polarity protein 1a |
| chr3_+_19299309 | 1.14 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr8_+_1189798 | 1.13 |
ENSDART00000193474
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr1_-_54765262 | 1.12 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr21_-_2238277 | 1.12 |
ENSDART00000165754
|
si:dkey-50i6.6
|
si:dkey-50i6.6 |
| chr1_-_55008882 | 1.11 |
ENSDART00000083572
|
zgc:136864
|
zgc:136864 |
| chr22_+_28337429 | 1.09 |
ENSDART00000166177
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr2_+_38556195 | 1.08 |
ENSDART00000138769
|
cdh24b
|
cadherin 24, type 2b |
| chr9_-_9282519 | 1.08 |
ENSDART00000146821
|
si:ch211-214p13.3
|
si:ch211-214p13.3 |
| chr21_-_41588129 | 1.07 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr25_-_29415369 | 1.05 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr11_-_40742424 | 1.05 |
ENSDART00000173399
ENSDART00000021369 |
tas1r3
|
taste receptor, type 1, member 3 |
| chr18_-_6460102 | 1.05 |
ENSDART00000137037
|
iqsec3b
|
IQ motif and Sec7 domain 3b |
| chr4_-_8152746 | 1.05 |
ENSDART00000012928
ENSDART00000177482 |
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr7_+_73295890 | 1.05 |
ENSDART00000174331
ENSDART00000174250 |
CABZ01083442.1
|
|
| chr21_+_5209476 | 1.04 |
ENSDART00000146400
|
st8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr12_+_5530247 | 1.03 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr9_-_8670158 | 1.03 |
ENSDART00000077296
|
col4a1
|
collagen, type IV, alpha 1 |
| chr2_-_10703621 | 1.02 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr2_-_38125657 | 0.97 |
ENSDART00000143433
|
cbln12
|
cerebellin 12 |
| chr2_-_37465517 | 0.96 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr16_+_16849220 | 0.92 |
ENSDART00000047409
ENSDART00000142155 |
myh14
|
myosin, heavy chain 14, non-muscle |
| chr5_+_28849155 | 0.92 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr4_-_67969695 | 0.91 |
ENSDART00000190016
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr6_+_41503854 | 0.91 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr2_+_38554260 | 0.91 |
ENSDART00000171527
|
cdh24b
|
cadherin 24, type 2b |
| chr19_-_20114149 | 0.90 |
ENSDART00000052620
|
npy
|
neuropeptide Y |
| chr15_-_2903475 | 0.89 |
ENSDART00000043226
|
guca1c
|
guanylate cyclase activator 1C |
| chr20_+_41664350 | 0.87 |
ENSDART00000186247
|
fam184a
|
family with sequence similarity 184, member A |
| chr19_-_47587719 | 0.87 |
ENSDART00000111108
|
CABZ01071972.1
|
|
| chr9_+_42157578 | 0.87 |
ENSDART00000142888
|
lrrc3
|
leucine rich repeat containing 3 |
| chr13_-_37631092 | 0.85 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr9_+_8851819 | 0.85 |
ENSDART00000160004
|
col4a2
|
collagen, type IV, alpha 2 |
| chr7_-_6425549 | 0.85 |
ENSDART00000173323
|
zgc:165555
|
zgc:165555 |
| chr14_-_2206476 | 0.84 |
ENSDART00000081870
|
pcdh2ab6
|
protocadherin 2 alpha b 6 |
| chr25_-_35137391 | 0.83 |
ENSDART00000182510
|
zgc:165555
|
zgc:165555 |
| chr10_-_30785501 | 0.83 |
ENSDART00000020054
|
opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr21_+_15870752 | 0.83 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr5_+_9360394 | 0.82 |
ENSDART00000124642
|
FP236810.2
|
|
| chr11_-_43226255 | 0.80 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr13_-_11644806 | 0.79 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr25_-_8160030 | 0.79 |
ENSDART00000067159
|
tph1a
|
tryptophan hydroxylase 1 (tryptophan 5-monooxygenase) a |
| chr20_+_6493648 | 0.78 |
ENSDART00000099846
|
mep1b
|
meprin A, beta |
| chr3_+_39656188 | 0.78 |
ENSDART00000186292
ENSDART00000156038 |
epn2
|
epsin 2 |
| chr16_+_22761846 | 0.77 |
ENSDART00000193028
|
CR854942.1
|
|
| chr20_-_16548912 | 0.77 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr23_-_1571682 | 0.76 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr7_+_14005111 | 0.76 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr19_-_6631900 | 0.75 |
ENSDART00000144571
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr19_-_6134802 | 0.75 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr20_-_40367493 | 0.75 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr14_-_2196267 | 0.74 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr11_+_29770966 | 0.73 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr22_+_1462177 | 0.73 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr22_+_37902976 | 0.73 |
ENSDART00000169327
|
kcnh1a
|
potassium voltage-gated channel, subfamily H (eag-related), member 1a |
| chr20_+_29586529 | 0.72 |
ENSDART00000178617
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr10_+_585719 | 0.72 |
ENSDART00000180167
|
smad4a
|
SMAD family member 4a |
| chr7_+_13039426 | 0.72 |
ENSDART00000171777
|
syt7b
|
synaptotagmin VIIb |
| chr16_-_27749172 | 0.71 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr8_+_11687254 | 0.70 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr13_+_33688474 | 0.70 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr11_+_43375326 | 0.70 |
ENSDART00000130131
|
sult6b1
|
sulfotransferase family, cytosolic, 6b, member 1 |
| chr13_-_2112008 | 0.69 |
ENSDART00000110814
|
fam83b
|
family with sequence similarity 83, member B |
| chr25_-_8030113 | 0.69 |
ENSDART00000104674
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr1_-_9195629 | 0.68 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr13_-_37608441 | 0.68 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr7_+_61764040 | 0.67 |
ENSDART00000056745
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr2_+_3428357 | 0.64 |
ENSDART00000125967
|
CU633991.1
|
|
| chr17_-_50331351 | 0.64 |
ENSDART00000149294
|
otofb
|
otoferlin b |
| chr21_-_22676323 | 0.64 |
ENSDART00000167392
|
gig2h
|
grass carp reovirus (GCRV)-induced gene 2h |
| chr6_+_40591149 | 0.63 |
ENSDART00000189060
ENSDART00000188298 |
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr21_+_26720803 | 0.63 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr18_-_7400075 | 0.62 |
ENSDART00000101250
|
si:dkey-30c15.13
|
si:dkey-30c15.13 |
| chr7_-_71837213 | 0.61 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr4_+_359970 | 0.61 |
ENSDART00000139832
|
tmem181
|
transmembrane protein 181 |
| chr13_-_47554194 | 0.61 |
ENSDART00000181508
|
acoxl
|
acyl-CoA oxidase-like |
| chr10_-_18468385 | 0.61 |
ENSDART00000183302
|
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr22_+_26804197 | 0.61 |
ENSDART00000135688
|
si:dkey-44g23.2
|
si:dkey-44g23.2 |
| chr1_-_46663997 | 0.61 |
ENSDART00000134450
|
ebpl
|
emopamil binding protein-like |
| chr1_+_10221568 | 0.61 |
ENSDART00000152424
|
npy2rl
|
neuropeptide Y receptor Y2, like |
| chr18_-_43880020 | 0.60 |
ENSDART00000185638
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr16_-_45398408 | 0.60 |
ENSDART00000004052
|
rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr5_+_25952340 | 0.60 |
ENSDART00000147188
|
trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr18_-_44888375 | 0.60 |
ENSDART00000160506
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr8_-_46700278 | 0.58 |
ENSDART00000143780
|
gpr153
|
G protein-coupled receptor 153 |
| chr12_-_28983584 | 0.57 |
ENSDART00000112374
|
zgc:171713
|
zgc:171713 |
| chr5_-_23118290 | 0.57 |
ENSDART00000132857
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr18_+_3634652 | 0.57 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr1_+_41454210 | 0.57 |
ENSDART00000148251
|
si:ch211-89o9.4
|
si:ch211-89o9.4 |
| chr14_-_27121854 | 0.57 |
ENSDART00000173119
|
pcdh11
|
protocadherin 11 |
| chr13_-_50234122 | 0.56 |
ENSDART00000164338
|
eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr9_+_51265283 | 0.56 |
ENSDART00000137426
|
gcgb
|
glucagon b |
| chr18_+_45228691 | 0.56 |
ENSDART00000127953
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr21_+_31811202 | 0.55 |
ENSDART00000189846
|
si:ch211-12m10.1
|
si:ch211-12m10.1 |
| chr23_-_30960506 | 0.55 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr22_-_17844117 | 0.55 |
ENSDART00000159363
|
BX908731.1
|
|
| chr25_-_21822426 | 0.55 |
ENSDART00000151993
|
zgc:158222
|
zgc:158222 |
| chr16_-_31933740 | 0.55 |
ENSDART00000125411
|
si:ch1073-90m23.1
|
si:ch1073-90m23.1 |
| chr20_-_25551676 | 0.55 |
ENSDART00000063081
|
cyp2ad3
|
cytochrome P450, family 2, subfamily AD, polypeptide 3 |
| chr20_+_4793790 | 0.54 |
ENSDART00000153486
|
lgals8a
|
galectin 8a |
| chr23_+_2185397 | 0.53 |
ENSDART00000109373
|
c1qtnf7
|
C1q and TNF related 7 |
| chr21_+_1378250 | 0.53 |
ENSDART00000186912
|
TCF4
|
transcription factor 4 |
| chr20_-_47188966 | 0.53 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr3_-_44059902 | 0.52 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr7_-_16204885 | 0.52 |
ENSDART00000171008
|
btr05
|
bloodthirsty-related gene family, member 5 |
| chr25_+_11281970 | 0.52 |
ENSDART00000180094
|
AKAP13
|
si:dkey-187e18.1 |
| chr21_+_26073104 | 0.51 |
ENSDART00000193273
|
rpl23a
|
ribosomal protein L23a |
| chr7_-_11596450 | 0.51 |
ENSDART00000173863
|
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr9_+_1313418 | 0.51 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr10_+_21722892 | 0.50 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr7_-_24373662 | 0.50 |
ENSDART00000173865
|
PTGR1 (1 of many)
|
si:dkey-11k2.7 |
| chr21_+_12010505 | 0.50 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
| chr19_+_32856907 | 0.50 |
ENSDART00000148232
|
rpl30
|
ribosomal protein L30 |
| chr9_-_23922011 | 0.49 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr5_-_55600689 | 0.48 |
ENSDART00000013229
|
gnaq
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr18_+_10689772 | 0.48 |
ENSDART00000126441
|
lepa
|
leptin a |
| chr1_-_56596701 | 0.48 |
ENSDART00000133693
|
LO018605.1
|
|
| chr15_-_23793641 | 0.48 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr25_+_37397031 | 0.48 |
ENSDART00000193643
ENSDART00000169132 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr7_+_5529967 | 0.47 |
ENSDART00000142709
|
si:dkeyp-67a8.4
|
si:dkeyp-67a8.4 |
| chr17_-_23631400 | 0.47 |
ENSDART00000079563
|
fas
|
Fas cell surface death receptor |
| chr14_-_6987649 | 0.47 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr12_-_26558038 | 0.46 |
ENSDART00000039510
|
tap2t
|
transporter associated with antigen processing, subunit type t, teleost specific |
| chr5_-_5035683 | 0.46 |
ENSDART00000170025
|
lmx1ba
|
LIM homeobox transcription factor 1, beta a |
| chr3_-_30888415 | 0.45 |
ENSDART00000124458
|
kmt5c
|
lysine methyltransferase 5C |
| chr24_-_1356668 | 0.45 |
ENSDART00000188935
|
nrp1a
|
neuropilin 1a |
| chr21_+_39361040 | 0.44 |
ENSDART00000085453
|
cluhb
|
clustered mitochondria (cluA/CLU1) homolog b |
| chr14_-_11507211 | 0.44 |
ENSDART00000186873
ENSDART00000109181 ENSDART00000186166 ENSDART00000186986 |
zgc:174917
|
zgc:174917 |
| chr16_-_41004731 | 0.44 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr11_+_36409457 | 0.44 |
ENSDART00000077641
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr24_+_37370064 | 0.44 |
ENSDART00000185870
|
si:ch211-183d21.3
|
si:ch211-183d21.3 |
| chr16_+_10329701 | 0.43 |
ENSDART00000172845
|
mdc1
|
mediator of DNA damage checkpoint 1 |
| chr13_-_37122217 | 0.43 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr21_-_2248239 | 0.43 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr19_-_35229336 | 0.43 |
ENSDART00000054274
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr22_-_11493236 | 0.43 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr1_-_47114310 | 0.42 |
ENSDART00000144899
ENSDART00000053157 |
setd4
|
SET domain containing 4 |
| chr8_+_28667021 | 0.42 |
ENSDART00000184292
|
slc13a3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr24_+_22039964 | 0.41 |
ENSDART00000081220
|
ankrd33ba
|
ankyrin repeat domain 33ba |
| chr1_+_11736420 | 0.41 |
ENSDART00000140329
|
chst12b
|
carbohydrate (chondroitin 4) sulfotransferase 12b |
| chr15_+_5321612 | 0.41 |
ENSDART00000174345
|
or113-1
|
odorant receptor, family A, subfamily 113, member 1 |
| chr7_-_19168375 | 0.41 |
ENSDART00000112447
|
il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr19_-_3821678 | 0.40 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr7_+_66739166 | 0.40 |
ENSDART00000154634
|
mrvi1
|
murine retrovirus integration site 1 homolog |
| chr9_+_38502524 | 0.39 |
ENSDART00000087229
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr18_+_6479963 | 0.39 |
ENSDART00000092752
ENSDART00000136333 |
wash1
|
WAS protein family homolog 1 |
| chr23_+_42292748 | 0.39 |
ENSDART00000166113
ENSDART00000158684 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr10_+_22134606 | 0.39 |
ENSDART00000155228
|
si:dkey-4c2.11
|
si:dkey-4c2.11 |
| chr14_+_23929059 | 0.39 |
ENSDART00000159582
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr16_-_17162843 | 0.39 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr15_-_2010926 | 0.38 |
ENSDART00000155688
|
dock10
|
dedicator of cytokinesis 10 |
| chr24_-_37472727 | 0.38 |
ENSDART00000134152
|
cluap1
|
clusterin associated protein 1 |
| chr1_-_40341306 | 0.37 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr11_+_45287541 | 0.37 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr9_+_29585943 | 0.37 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr8_+_47099033 | 0.36 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr1_-_40208237 | 0.36 |
ENSDART00000191987
|
AL953865.1
|
|
| chr9_+_12948511 | 0.36 |
ENSDART00000135797
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr21_-_43398457 | 0.36 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr18_+_18863167 | 0.36 |
ENSDART00000091094
|
pllp
|
plasmolipin |
Network of associatons between targets according to the STRING database.
First level regulatory network of bcl6aa+bcl6ab+bcl6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.5 | 1.9 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.4 | 1.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.4 | 2.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.4 | 2.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.3 | 1.3 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.3 | 1.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 2.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 1.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.2 | 3.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 0.9 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.2 | 0.6 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 1.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.2 | 0.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.4 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.7 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.5 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.1 | 0.5 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 0.3 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 0.3 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 0.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.6 | GO:0050951 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.1 | 2.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.3 | GO:0002858 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.1 | 0.4 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 1.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.6 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.3 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.1 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.3 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.1 | 0.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.4 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.0 | GO:2000651 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 1.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.6 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 2.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.0 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.5 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.6 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.5 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.2 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.8 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.7 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 1.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.2 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.2 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.5 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.2 | GO:0050688 | regulation of response to biotic stimulus(GO:0002831) regulation of defense response to virus(GO:0050688) |
| 0.0 | 1.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.1 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.0 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 2.1 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 1.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 2.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0001508 | action potential(GO:0001508) |
| 0.0 | 0.3 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 1.4 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.3 | 2.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 0.7 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 0.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.6 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.5 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.8 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 0.4 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 1.0 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 4.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 2.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 2.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.3 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 2.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.3 | 2.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 1.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.3 | 1.1 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.3 | 1.9 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.3 | 1.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 1.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 0.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 0.9 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 0.7 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 1.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 0.5 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.1 | 0.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 1.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.5 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 1.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.6 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.9 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 2.4 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.4 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.0 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.5 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.6 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.8 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 0.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 0.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.2 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 2.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.9 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.6 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |