Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for bcl11aa
Z-value: 0.67
Transcription factors associated with bcl11aa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
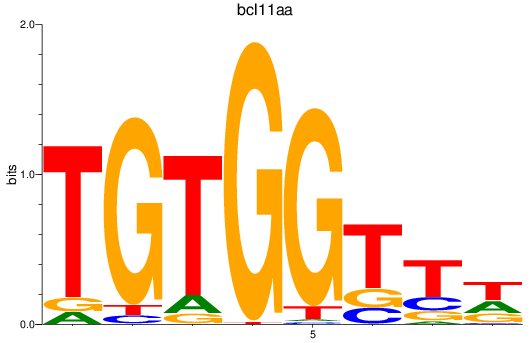
bcl11aa
|
ENSDARG00000061352 | BAF chromatin remodeling complex subunit BCL11A a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bcl11aa | dr11_v1_chr13_+_25846528_25846528 | 0.52 | 2.3e-02 | Click! |
Activity profile of bcl11aa motif
Sorted Z-values of bcl11aa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_18974064 | 2.41 |
ENSDART00000079248
|
slc6a19b
|
solute carrier family 6 (neutral amino acid transporter), member 19b |
| chr22_-_31517300 | 1.49 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr16_-_13612650 | 1.43 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr9_-_44939104 | 1.38 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr5_-_18046053 | 1.37 |
ENSDART00000144898
|
rnf215
|
ring finger protein 215 |
| chr15_+_39977461 | 1.33 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr18_+_8805575 | 1.20 |
ENSDART00000137098
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr5_-_38384289 | 1.08 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr2_+_22694382 | 1.04 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr7_+_49865049 | 0.99 |
ENSDART00000164165
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr10_+_38526496 | 0.96 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr21_-_21096437 | 0.93 |
ENSDART00000186552
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr23_+_40908583 | 0.89 |
ENSDART00000180933
|
LO017845.1
|
|
| chr17_+_15882533 | 0.86 |
ENSDART00000164124
|
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr24_+_37722048 | 0.85 |
ENSDART00000191918
|
rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr5_-_43682930 | 0.83 |
ENSDART00000075017
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr8_-_43997538 | 0.83 |
ENSDART00000186449
|
rimbp2
|
RIMS binding protein 2 |
| chr25_+_4541211 | 0.82 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr4_+_20255160 | 0.82 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr8_-_43456025 | 0.81 |
ENSDART00000001092
ENSDART00000140618 |
ncor2
|
nuclear receptor corepressor 2 |
| chr24_+_26379441 | 0.80 |
ENSDART00000137786
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr13_-_23270576 | 0.78 |
ENSDART00000132828
|
si:dkey-103j14.5
|
si:dkey-103j14.5 |
| chr5_-_18446483 | 0.78 |
ENSDART00000180027
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr11_+_38280454 | 0.78 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr19_-_22488952 | 0.77 |
ENSDART00000179856
ENSDART00000141503 |
pleca
|
plectin a |
| chr13_+_31177934 | 0.73 |
ENSDART00000144568
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr15_-_26636826 | 0.73 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr14_+_29975268 | 0.73 |
ENSDART00000172985
|
cyp4v7
|
cytochrome P450, family 4, subfamily V, polypeptide 7 |
| chr11_+_30058139 | 0.70 |
ENSDART00000112254
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr3_-_45298487 | 0.69 |
ENSDART00000102245
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr7_-_2163361 | 0.69 |
ENSDART00000173654
|
si:cabz01007812.1
|
si:cabz01007812.1 |
| chr3_-_37476278 | 0.68 |
ENSDART00000083442
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr6_+_10534752 | 0.67 |
ENSDART00000090874
|
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr21_+_27283054 | 0.67 |
ENSDART00000139058
|
si:dkey-175m17.7
|
si:dkey-175m17.7 |
| chr2_-_5505094 | 0.67 |
ENSDART00000145035
|
saga
|
S-antigen; retina and pineal gland (arrestin) a |
| chr9_+_17984358 | 0.65 |
ENSDART00000192399
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr2_+_38804223 | 0.61 |
ENSDART00000147939
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr5_+_61556172 | 0.61 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr9_+_41612642 | 0.61 |
ENSDART00000138473
|
spegb
|
SPEG complex locus b |
| chr21_+_5915041 | 0.60 |
ENSDART00000151370
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr22_-_38819603 | 0.59 |
ENSDART00000104437
|
si:ch211-262h13.5
|
si:ch211-262h13.5 |
| chr3_-_56512473 | 0.59 |
ENSDART00000125439
ENSDART00000181743 |
cyth1a
|
cytohesin 1a |
| chr3_+_22905341 | 0.58 |
ENSDART00000111435
|
hdac5
|
histone deacetylase 5 |
| chr3_+_15773991 | 0.56 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr17_+_19730001 | 0.53 |
ENSDART00000155025
|
grem2a
|
gremlin 2, DAN family BMP antagonist a |
| chr11_-_40257225 | 0.52 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
| chr2_-_24369087 | 0.51 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr22_-_20379045 | 0.51 |
ENSDART00000183511
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr10_+_29204581 | 0.50 |
ENSDART00000148503
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr7_+_39706004 | 0.50 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr16_+_27957808 | 0.48 |
ENSDART00000133696
|
znf804b
|
zinc finger protein 804B |
| chr16_+_8716800 | 0.47 |
ENSDART00000124693
ENSDART00000181961 |
cabz01093075.1
|
cabz01093075.1 |
| chr21_+_37357578 | 0.46 |
ENSDART00000143621
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr14_-_33177935 | 0.46 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr17_+_27134806 | 0.46 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr17_-_44776224 | 0.44 |
ENSDART00000156195
|
zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr8_+_47188154 | 0.44 |
ENSDART00000137319
|
si:dkeyp-100a1.6
|
si:dkeyp-100a1.6 |
| chr7_+_50109239 | 0.44 |
ENSDART00000021605
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr20_+_46216431 | 0.40 |
ENSDART00000185384
|
stx7l
|
syntaxin 7-like |
| chr15_-_36352983 | 0.40 |
ENSDART00000059786
|
si:dkey-23k10.5
|
si:dkey-23k10.5 |
| chr3_-_56924654 | 0.40 |
ENSDART00000157038
|
hid1a
|
HID1 domain containing a |
| chr14_-_45702721 | 0.38 |
ENSDART00000165727
|
si:dkey-226n24.1
|
si:dkey-226n24.1 |
| chr4_+_34126849 | 0.38 |
ENSDART00000162442
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr8_+_36803415 | 0.38 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr11_+_23957440 | 0.37 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr17_-_45254585 | 0.37 |
ENSDART00000185507
ENSDART00000172080 |
ttbk2a
|
tau tubulin kinase 2a |
| chr3_-_31833266 | 0.37 |
ENSDART00000084932
ENSDART00000183617 |
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr15_-_38154616 | 0.37 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr18_+_40354998 | 0.36 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr25_-_27564590 | 0.36 |
ENSDART00000073508
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr6_-_21830405 | 0.36 |
ENSDART00000151803
ENSDART00000113497 |
setd5
|
SET domain containing 5 |
| chr6_+_45918981 | 0.35 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr2_-_14387335 | 0.35 |
ENSDART00000189332
ENSDART00000164786 ENSDART00000188572 |
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr17_+_8899016 | 0.34 |
ENSDART00000150214
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr24_-_31306724 | 0.34 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr9_-_22352280 | 0.33 |
ENSDART00000115384
ENSDART00000060361 |
crygm5
|
crystallin, gamma M5 |
| chr2_+_3516913 | 0.32 |
ENSDART00000109346
|
CU693445.1
|
|
| chr10_+_19039507 | 0.31 |
ENSDART00000193538
ENSDART00000111952 ENSDART00000180093 |
igsf9a
|
immunoglobulin superfamily, member 9a |
| chr8_+_10835456 | 0.30 |
ENSDART00000151388
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr10_-_15128771 | 0.30 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr13_-_47403154 | 0.29 |
ENSDART00000114318
|
bcl2l11
|
BCL2 like 11 |
| chr24_-_18876877 | 0.28 |
ENSDART00000186269
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr4_+_3980247 | 0.28 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr9_-_30160897 | 0.28 |
ENSDART00000143986
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr11_+_4026229 | 0.28 |
ENSDART00000041417
|
camk1b
|
calcium/calmodulin-dependent protein kinase Ib |
| chr24_+_10302955 | 0.27 |
ENSDART00000112455
ENSDART00000066794 ENSDART00000149432 |
otulina
otulina
|
OTU deubiquitinase with linear linkage specificity a OTU deubiquitinase with linear linkage specificity a |
| chr19_+_7552699 | 0.26 |
ENSDART00000180788
ENSDART00000115058 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr24_-_36680261 | 0.26 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr1_+_23406403 | 0.26 |
ENSDART00000189921
|
chrna9
|
cholinergic receptor, nicotinic, alpha 9 |
| chr4_+_17336557 | 0.25 |
ENSDART00000111650
|
pmch
|
pro-melanin-concentrating hormone |
| chr6_-_7438584 | 0.25 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr2_+_24786765 | 0.24 |
ENSDART00000141030
|
pde4ca
|
phosphodiesterase 4C, cAMP-specific a |
| chr16_+_38820660 | 0.24 |
ENSDART00000182538
ENSDART00000189879 |
trhra
|
thyrotropin-releasing hormone receptor a |
| chr18_+_40525408 | 0.23 |
ENSDART00000132833
|
vwa7
|
von Willebrand factor A domain containing 7 |
| chr3_-_25369557 | 0.23 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr9_+_33267211 | 0.21 |
ENSDART00000025635
|
usp9
|
ubiquitin specific peptidase 9 |
| chr6_-_19023468 | 0.21 |
ENSDART00000184729
|
sept9b
|
septin 9b |
| chr9_+_51655636 | 0.21 |
ENSDART00000169908
|
RBMS1 (1 of many)
|
RNA binding motif single stranded interacting protein 1 |
| chr21_-_28439596 | 0.20 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr11_-_12800945 | 0.20 |
ENSDART00000191178
|
txlng
|
taxilin gamma |
| chr1_+_39597809 | 0.20 |
ENSDART00000122700
|
tenm3
|
teneurin transmembrane protein 3 |
| chr5_+_54422941 | 0.20 |
ENSDART00000175157
|
traf2b
|
Tnf receptor-associated factor 2b |
| chr16_+_26439939 | 0.19 |
ENSDART00000143073
|
trim35-28
|
tripartite motif containing 35-28 |
| chr11_-_12801157 | 0.19 |
ENSDART00000103449
|
txlng
|
taxilin gamma |
| chr19_-_8798178 | 0.19 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr3_+_51660158 | 0.19 |
ENSDART00000155512
ENSDART00000171764 |
chmp6a
|
charged multivesicular body protein 6a |
| chr16_-_16619854 | 0.17 |
ENSDART00000150512
ENSDART00000191306 ENSDART00000181773 ENSDART00000183231 |
cyp21a2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr2_+_21855291 | 0.17 |
ENSDART00000186204
|
ca8
|
carbonic anhydrase VIII |
| chr16_+_10963602 | 0.15 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr7_+_46368520 | 0.15 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr25_-_22889519 | 0.15 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr25_-_27564205 | 0.14 |
ENSDART00000157319
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr20_+_38285671 | 0.14 |
ENSDART00000061432
|
ccl38a.4
|
chemokine (C-C motif) ligand 38, duplicate 4 |
| chr19_-_22478888 | 0.14 |
ENSDART00000090679
|
pleca
|
plectin a |
| chr23_-_4975452 | 0.13 |
ENSDART00000105241
ENSDART00000169978 |
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr2_-_36925561 | 0.13 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr11_-_29685853 | 0.13 |
ENSDART00000128468
|
nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr17_+_5351922 | 0.13 |
ENSDART00000105394
|
runx2a
|
runt-related transcription factor 2a |
| chr17_+_5426087 | 0.13 |
ENSDART00000131430
ENSDART00000188636 |
runx2a
|
runt-related transcription factor 2a |
| chr20_+_40150612 | 0.13 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr25_+_18953756 | 0.12 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr15_+_29662401 | 0.12 |
ENSDART00000135540
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr7_-_29571615 | 0.12 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr16_+_19014886 | 0.11 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr11_-_30601401 | 0.11 |
ENSDART00000172244
ENSDART00000046800 |
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr25_+_15997957 | 0.11 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr20_-_14054083 | 0.11 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr24_+_39277043 | 0.11 |
ENSDART00000165458
|
MPRIP
|
si:ch73-103b11.2 |
| chr6_+_13606410 | 0.11 |
ENSDART00000104716
|
asic4b
|
acid-sensing (proton-gated) ion channel family member 4b |
| chr16_-_39900665 | 0.10 |
ENSDART00000136719
|
rbms3
|
RNA binding motif, single stranded interacting protein |
| chr6_-_16394528 | 0.10 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr16_+_8695595 | 0.10 |
ENSDART00000173249
|
si:cabz01093077.1
|
si:cabz01093077.1 |
| chr24_+_13635387 | 0.10 |
ENSDART00000105997
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr22_-_10470663 | 0.09 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr8_+_45294767 | 0.09 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr20_+_52554352 | 0.09 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr5_-_60885935 | 0.09 |
ENSDART00000128350
|
rad51d
|
RAD51 paralog D |
| chr16_+_11724230 | 0.09 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr14_+_24845941 | 0.08 |
ENSDART00000187513
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr21_-_11632403 | 0.08 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr19_+_3849378 | 0.08 |
ENSDART00000166218
ENSDART00000159228 |
oscp1a
|
organic solute carrier partner 1a |
| chr25_+_31929325 | 0.07 |
ENSDART00000181095
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr14_-_33095917 | 0.07 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr7_-_7810348 | 0.07 |
ENSDART00000171984
|
cxcl19
|
chemokine (C-X-C motif) ligand 19 |
| chr7_-_7984015 | 0.07 |
ENSDART00000163203
|
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr5_-_31926906 | 0.06 |
ENSDART00000187340
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr25_-_35996141 | 0.06 |
ENSDART00000149074
|
sall1b
|
spalt-like transcription factor 1b |
| chr6_+_34868156 | 0.05 |
ENSDART00000149364
|
il23r
|
interleukin 23 receptor |
| chr17_-_26868169 | 0.05 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr22_-_10397600 | 0.05 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr5_+_24156170 | 0.05 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr24_-_18877118 | 0.05 |
ENSDART00000092783
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr11_+_30057762 | 0.04 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr1_+_33217911 | 0.03 |
ENSDART00000136761
|
prkx
|
protein kinase, X-linked |
| chr8_-_25690966 | 0.03 |
ENSDART00000033701
|
sema3ga
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ga |
| chr6_+_38626926 | 0.03 |
ENSDART00000190339
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr16_-_29527412 | 0.03 |
ENSDART00000175376
|
onecutl
|
one cut domain, family member, like |
| chr20_-_20821783 | 0.02 |
ENSDART00000152577
ENSDART00000027603 ENSDART00000145601 |
ckbb
|
creatine kinase, brain b |
| chr14_+_45970033 | 0.02 |
ENSDART00000047716
|
fermt3b
|
fermitin family member 3b |
| chr2_-_6482240 | 0.01 |
ENSDART00000132623
|
rgs13
|
regulator of G protein signaling 13 |
| chr16_+_26439518 | 0.01 |
ENSDART00000041787
|
trim35-28
|
tripartite motif containing 35-28 |
| chr7_+_59677273 | 0.00 |
ENSDART00000039535
ENSDART00000132044 |
trmt44
|
tRNA methyltransferase 44 homolog |
Network of associatons between targets according to the STRING database.
First level regulatory network of bcl11aa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.2 | 0.7 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 0.8 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 0.8 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.4 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.6 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 1.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.8 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.5 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 1.0 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 1.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.2 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.7 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 1.5 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.8 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.3 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 1.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.1 | GO:0008584 | male gonad development(GO:0008584) |
| 0.0 | 1.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 2.6 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.4 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.6 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.6 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 1.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 1.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 0.8 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 0.6 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.7 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 1.5 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.6 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.9 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.5 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 1.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 3.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.8 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |