Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
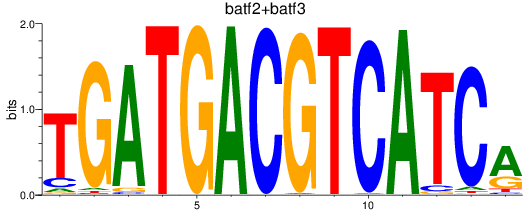
Results for batf2+batf3
Z-value: 0.66
Transcription factors associated with batf2+batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
batf3
|
ENSDARG00000042577 | basic leucine zipper transcription factor, ATF-like 3 |
|
batf2
|
ENSDARG00000105562 | si_dkey-23i12.7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| batf3 | dr11_v1_chr20_-_37813863_37813863 | -0.14 | 5.8e-01 | Click! |
| si:dkey-23i12.7 | dr11_v1_chr7_+_25049545_25049577 | 0.07 | 7.6e-01 | Click! |
Activity profile of batf2+batf3 motif
Sorted Z-values of batf2+batf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_23308286 | 2.33 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr22_+_1911269 | 2.05 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr21_-_23307653 | 1.91 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr9_-_33121535 | 1.63 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr22_+_1668471 | 1.21 |
ENSDART00000163358
|
si:dkey-1b17.5
|
si:dkey-1b17.5 |
| chr22_+_1947494 | 1.18 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr22_+_2183110 | 1.11 |
ENSDART00000159279
ENSDART00000121703 |
znf1152
|
zinc finger protein 1152 |
| chr4_+_30775376 | 1.03 |
ENSDART00000158528
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr22_+_2819613 | 1.02 |
ENSDART00000131234
|
si:dkey-20i20.3
|
si:dkey-20i20.3 |
| chr22_+_2715815 | 0.99 |
ENSDART00000042770
|
znf1163
|
zinc finger protein 1163 |
| chr3_-_16413606 | 0.99 |
ENSDART00000127309
ENSDART00000017172 ENSDART00000136465 |
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr25_-_6447835 | 0.97 |
ENSDART00000012820
|
snupn
|
snurportin 1 |
| chr16_-_29387215 | 0.84 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr22_+_2228919 | 0.82 |
ENSDART00000133475
|
znf1161
|
zinc finger protein 1161 |
| chr22_+_2403068 | 0.80 |
ENSDART00000132925
ENSDART00000132569 |
zgc:112977
|
zgc:112977 |
| chr22_+_1821718 | 0.79 |
ENSDART00000132220
|
znf1002
|
zinc finger protein 1002 |
| chr3_-_40162843 | 0.79 |
ENSDART00000129664
ENSDART00000025285 |
drg2
|
developmentally regulated GTP binding protein 2 |
| chr4_-_61651223 | 0.78 |
ENSDART00000172688
|
si:dkey-26i24.1
|
si:dkey-26i24.1 |
| chr21_-_14762944 | 0.77 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr23_-_40766518 | 0.74 |
ENSDART00000127420
|
si:dkeyp-27c8.2
|
si:dkeyp-27c8.2 |
| chr5_-_19014589 | 0.73 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr15_+_34963316 | 0.68 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr4_-_61650771 | 0.67 |
ENSDART00000168593
|
si:dkey-26i24.1
|
si:dkey-26i24.1 |
| chr1_-_20068155 | 0.64 |
ENSDART00000102993
|
mettl14
|
methyltransferase like 14 |
| chr22_+_1641547 | 0.63 |
ENSDART00000159804
|
si:dkey-1b17.2
|
si:dkey-1b17.2 |
| chr22_+_2524615 | 0.62 |
ENSDART00000134277
|
znf1003
|
zinc finger protein 1003 |
| chr4_+_70556298 | 0.58 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr16_+_27614989 | 0.57 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr25_-_6448050 | 0.56 |
ENSDART00000180616
|
snupn
|
snurportin 1 |
| chr7_+_8670398 | 0.54 |
ENSDART00000164195
|
si:ch211-1o7.2
|
si:ch211-1o7.2 |
| chr22_+_1940595 | 0.51 |
ENSDART00000163506
|
znf1167
|
zinc finger protein 1167 |
| chr22_+_1930589 | 0.48 |
ENSDART00000159807
|
znf1153
|
zinc finger protein 1153 |
| chr22_+_2751887 | 0.48 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr22_+_1922293 | 0.45 |
ENSDART00000165758
|
znf1181
|
zinc finger protein 1181 |
| chr22_+_1751640 | 0.44 |
ENSDART00000162093
|
znf1169
|
zinc finger protein 1169 |
| chr14_-_32959851 | 0.44 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr3_-_49514874 | 0.43 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr3_-_40658820 | 0.43 |
ENSDART00000191948
|
rnf216
|
ring finger protein 216 |
| chr8_-_21110233 | 0.42 |
ENSDART00000127371
ENSDART00000100276 |
tmco1
|
transmembrane and coiled-coil domains 1 |
| chr22_+_1953096 | 0.41 |
ENSDART00000166630
ENSDART00000186234 |
si:dkey-15h8.16
|
si:dkey-15h8.16 |
| chr22_+_1853999 | 0.38 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr22_+_2345752 | 0.37 |
ENSDART00000143687
|
zgc:174224
|
zgc:174224 |
| chr20_-_21994901 | 0.37 |
ENSDART00000004984
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr5_+_20823409 | 0.36 |
ENSDART00000093185
ENSDART00000142894 |
limk2
|
LIM domain kinase 2 |
| chr10_+_28306749 | 0.35 |
ENSDART00000142016
|
ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr22_+_2801464 | 0.34 |
ENSDART00000132419
|
si:dkey-20i20.7
|
si:dkey-20i20.7 |
| chr22_+_2111120 | 0.33 |
ENSDART00000165525
|
znf1144
|
zinc finger protein 1144 |
| chr22_+_2298419 | 0.31 |
ENSDART00000143499
|
znf1180
|
zinc finger protein 1180 |
| chr5_+_11812089 | 0.30 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr17_+_32500387 | 0.26 |
ENSDART00000018423
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr18_-_12451772 | 0.25 |
ENSDART00000175083
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr13_+_30225262 | 0.25 |
ENSDART00000141945
|
rps24
|
ribosomal protein S24 |
| chr5_+_37035978 | 0.24 |
ENSDART00000167418
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr10_-_20523405 | 0.23 |
ENSDART00000114824
|
ddhd2
|
DDHD domain containing 2 |
| chr22_+_1501566 | 0.23 |
ENSDART00000167788
|
AL954741.3
|
|
| chr4_-_25271455 | 0.21 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr4_+_63484571 | 0.20 |
ENSDART00000169518
ENSDART00000168681 |
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr4_+_30785713 | 0.19 |
ENSDART00000165945
|
si:dkey-178j11.5
|
si:dkey-178j11.5 |
| chr22_+_30330574 | 0.17 |
ENSDART00000104751
|
mxi1
|
max interactor 1, dimerization protein |
| chr11_-_43226255 | 0.17 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr16_-_54942532 | 0.15 |
ENSDART00000078887
ENSDART00000101402 |
tmem222a
|
transmembrane protein 222a |
| chr22_+_2844865 | 0.14 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr16_-_41667101 | 0.13 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr7_-_8156169 | 0.11 |
ENSDART00000182960
ENSDART00000190184 ENSDART00000190885 ENSDART00000189867 ENSDART00000160836 |
si:cabz01030277.1
si:ch211-163c2.3
|
si:cabz01030277.1 si:ch211-163c2.3 |
| chr7_+_40228422 | 0.08 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr22_+_1615691 | 0.05 |
ENSDART00000171555
|
si:ch211-255f4.13
|
si:ch211-255f4.13 |
| chr22_+_1708760 | 0.02 |
ENSDART00000170471
|
si:dkey-1b17.9
|
si:dkey-1b17.9 |
| chr17_+_15674052 | 0.01 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr22_+_2189356 | 0.01 |
ENSDART00000157502
ENSDART00000164188 |
znf1165
|
zinc finger protein 1165 |
| chr7_+_29301497 | 0.00 |
ENSDART00000099327
|
rab8b
|
RAB8B, member RAS oncogene family |
| chr22_+_2390544 | 0.00 |
ENSDART00000147345
ENSDART00000133611 |
zgc:171435
|
zgc:171435 |
Network of associatons between targets according to the STRING database.
First level regulatory network of batf2+batf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.3 | 4.2 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.7 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 0.6 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.0 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.2 | 0.6 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.2 | 0.8 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 4.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.7 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |