Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
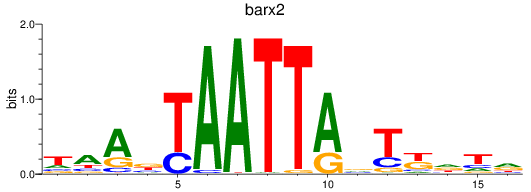
Results for barx2
Z-value: 1.55
Transcription factors associated with barx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barx2
|
ENSDARG00000041098 | BARX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barx2 | dr11_v1_chr18_+_47313715_47313715 | 0.47 | 4.4e-02 | Click! |
Activity profile of barx2 motif
Sorted Z-values of barx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_46313082 | 3.30 |
ENSDART00000153830
|
si:ch1073-190k2.1
|
si:ch1073-190k2.1 |
| chr8_+_23521974 | 2.90 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr11_+_24002503 | 2.48 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr23_-_21446985 | 2.35 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr1_+_52662203 | 2.20 |
ENSDART00000141530
|
osbp
|
oxysterol binding protein |
| chr18_-_48983690 | 2.16 |
ENSDART00000182359
|
FO681288.3
|
|
| chr18_+_3579829 | 2.15 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr16_+_28994709 | 2.11 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr23_+_40460333 | 2.06 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr25_+_37126921 | 1.99 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr4_+_2482046 | 1.90 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr6_-_12588044 | 1.82 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr24_-_29586082 | 1.82 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr20_-_47188966 | 1.73 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr3_-_12026741 | 1.63 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr21_-_2814709 | 1.63 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr5_+_71802014 | 1.59 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr1_-_19502322 | 1.53 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr8_+_6576940 | 1.50 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr8_+_26141680 | 1.35 |
ENSDART00000078334
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr20_+_41664350 | 1.34 |
ENSDART00000186247
|
fam184a
|
family with sequence similarity 184, member A |
| chr23_-_900795 | 1.34 |
ENSDART00000190517
ENSDART00000182849 ENSDART00000111456 ENSDART00000185430 |
rbm10
|
RNA binding motif protein 10 |
| chr3_-_40054615 | 1.31 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr12_-_7854216 | 1.30 |
ENSDART00000149594
|
ank3b
|
ankyrin 3b |
| chr3_-_59297532 | 1.28 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr1_-_2457546 | 1.27 |
ENSDART00000103795
|
ggact.1
|
gamma-glutamylamine cyclotransferase, tandem duplicate 1 |
| chr9_+_2574122 | 1.27 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr2_-_17694504 | 1.25 |
ENSDART00000133709
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr21_-_36571804 | 1.24 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr3_+_35542067 | 1.20 |
ENSDART00000146529
ENSDART00000084549 |
rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr19_-_42045372 | 1.19 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr20_-_53078607 | 1.17 |
ENSDART00000163494
ENSDART00000191730 |
CABZ01066813.1
|
|
| chr23_+_7710721 | 1.17 |
ENSDART00000186852
ENSDART00000161193 |
kif3b
|
kinesin family member 3B |
| chr7_-_25895189 | 1.17 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr11_-_6974022 | 1.16 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr12_+_4971515 | 1.16 |
ENSDART00000161076
|
arhgap27
|
Rho GTPase activating protein 27 |
| chr3_+_62339264 | 1.14 |
ENSDART00000174569
|
BX470259.3
|
|
| chr9_+_7998794 | 1.14 |
ENSDART00000138167
|
myo16
|
myosin XVI |
| chr8_-_50888806 | 1.14 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr11_+_45436703 | 1.12 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr20_-_9095105 | 1.10 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr4_-_2219705 | 1.09 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr5_-_66702479 | 1.09 |
ENSDART00000129197
|
mn1b
|
meningioma 1b |
| chr22_+_2769236 | 1.07 |
ENSDART00000141836
|
si:dkey-20i20.10
|
si:dkey-20i20.10 |
| chr9_+_24065855 | 1.07 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr15_-_40157513 | 1.07 |
ENSDART00000184014
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr1_+_54124209 | 1.07 |
ENSDART00000187730
|
LO017722.1
|
|
| chr15_-_40157331 | 1.07 |
ENSDART00000187958
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr10_-_33297864 | 1.06 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr19_+_1688727 | 1.05 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr1_-_47071979 | 1.05 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr21_-_13972745 | 1.04 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr19_-_26869103 | 1.03 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr23_+_2789422 | 1.03 |
ENSDART00000156954
|
plcg1
|
phospholipase C, gamma 1 |
| chr12_-_48006835 | 1.03 |
ENSDART00000108989
|
adamts14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
| chr22_-_21897203 | 1.01 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr16_+_54209504 | 0.99 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr15_-_40267485 | 0.98 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr25_+_35891342 | 0.98 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr5_-_51198430 | 0.97 |
ENSDART00000132503
ENSDART00000097473 ENSDART00000165870 |
snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr10_+_45089820 | 0.96 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr22_-_37797695 | 0.95 |
ENSDART00000085931
ENSDART00000185443 |
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr11_+_29975830 | 0.95 |
ENSDART00000148929
|
si:ch73-226l13.2
|
si:ch73-226l13.2 |
| chr17_-_16422654 | 0.92 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr25_+_16116740 | 0.92 |
ENSDART00000139778
|
far1
|
fatty acyl CoA reductase 1 |
| chr8_+_25767610 | 0.92 |
ENSDART00000062406
|
cacna1sb
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, b |
| chr7_-_31922432 | 0.92 |
ENSDART00000188398
|
lin7c
|
lin-7 homolog C (C. elegans) |
| chr7_+_24573721 | 0.91 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr17_-_200316 | 0.91 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr23_-_26521970 | 0.91 |
ENSDART00000143712
|
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr8_+_7097929 | 0.90 |
ENSDART00000188955
ENSDART00000184772 ENSDART00000109581 |
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr5_+_19933356 | 0.90 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr13_+_646700 | 0.89 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr9_+_21277846 | 0.89 |
ENSDART00000139620
ENSDART00000110996 ENSDART00000111899 |
lats2
|
large tumor suppressor kinase 2 |
| chr1_-_56080112 | 0.88 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr18_+_50461981 | 0.87 |
ENSDART00000158761
|
CU896640.1
|
|
| chr13_+_24750078 | 0.87 |
ENSDART00000021053
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr24_+_7336807 | 0.86 |
ENSDART00000137010
|
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr15_-_40157165 | 0.86 |
ENSDART00000192991
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr12_-_35885349 | 0.83 |
ENSDART00000085662
|
cep131
|
centrosomal protein 131 |
| chr7_+_26534131 | 0.83 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr8_-_19246342 | 0.83 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr7_+_1530024 | 0.82 |
ENSDART00000163082
|
tox4b
|
TOX high mobility group box family member 4 b |
| chr10_-_21054059 | 0.82 |
ENSDART00000139733
|
pcdh1a
|
protocadherin 1a |
| chr19_+_19775757 | 0.81 |
ENSDART00000164677
|
hoxa3a
|
homeobox A3a |
| chr1_+_11977426 | 0.80 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr17_+_49281597 | 0.80 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr19_-_205104 | 0.80 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr16_+_11623956 | 0.80 |
ENSDART00000137788
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr20_-_35578435 | 0.78 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr2_-_58142854 | 0.78 |
ENSDART00000169909
|
CABZ01110881.1
|
|
| chr1_-_5455498 | 0.78 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr25_-_19420949 | 0.77 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr17_+_50524573 | 0.76 |
ENSDART00000187974
|
CR382385.1
|
|
| chr23_+_45785563 | 0.76 |
ENSDART00000186027
|
CABZ01088036.1
|
|
| chr15_-_14552101 | 0.76 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr15_-_47115787 | 0.76 |
ENSDART00000192601
|
CABZ01079081.1
|
|
| chr9_+_48761455 | 0.76 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr1_+_39482588 | 0.75 |
ENSDART00000181790
|
tenm3
|
teneurin transmembrane protein 3 |
| chr7_-_71829649 | 0.75 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr22_+_18816662 | 0.75 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr1_+_51312752 | 0.74 |
ENSDART00000063938
|
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr11_+_29770966 | 0.74 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr22_-_37796998 | 0.74 |
ENSDART00000124742
ENSDART00000191232 |
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr20_-_39333657 | 0.73 |
ENSDART00000153720
ENSDART00000142164 |
ccl38.1
|
chemokine (C-C motif) ligand 38, duplicate 1 |
| chr19_+_40069524 | 0.73 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr5_+_63668735 | 0.73 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr7_+_53156810 | 0.73 |
ENSDART00000189816
|
cdh29
|
cadherin 29 |
| chr2_-_3678029 | 0.72 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr10_-_2524917 | 0.72 |
ENSDART00000188642
|
CU856539.1
|
|
| chr17_-_20717845 | 0.72 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr23_-_18024543 | 0.71 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr22_-_17844117 | 0.71 |
ENSDART00000159363
|
BX908731.1
|
|
| chr23_-_33709964 | 0.70 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr10_-_37072776 | 0.70 |
ENSDART00000110835
|
myo18aa
|
myosin XVIIIAa |
| chr23_-_1660708 | 0.69 |
ENSDART00000175138
|
CU693481.1
|
|
| chr21_+_21195487 | 0.69 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr23_-_1017605 | 0.69 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr24_+_23840821 | 0.69 |
ENSDART00000128595
ENSDART00000127188 |
pola1
|
polymerase (DNA directed), alpha 1 |
| chr21_-_44081540 | 0.69 |
ENSDART00000130833
|
FO704810.1
|
|
| chr6_+_612594 | 0.69 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr11_-_40742424 | 0.68 |
ENSDART00000173399
ENSDART00000021369 |
tas1r3
|
taste receptor, type 1, member 3 |
| chr19_-_6239248 | 0.68 |
ENSDART00000014127
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr12_+_48480632 | 0.68 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr12_-_314899 | 0.68 |
ENSDART00000066579
|
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr24_+_36204028 | 0.68 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr24_+_7336411 | 0.68 |
ENSDART00000191170
|
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr17_-_29224908 | 0.68 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr8_+_52637507 | 0.67 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr21_-_2232640 | 0.67 |
ENSDART00000157754
|
zgc:113343
|
zgc:113343 |
| chr3_-_1989000 | 0.67 |
ENSDART00000193936
|
BX005442.2
|
|
| chr8_-_7567815 | 0.67 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr1_+_6862917 | 0.67 |
ENSDART00000182953
|
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr14_-_30387894 | 0.66 |
ENSDART00000176136
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr19_+_31873308 | 0.65 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr2_-_59205393 | 0.65 |
ENSDART00000056417
ENSDART00000182452 ENSDART00000141876 |
ftr30
|
finTRIM family, member 30 |
| chr17_-_4245902 | 0.64 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr9_-_32604414 | 0.64 |
ENSDART00000088876
ENSDART00000166502 |
satb2
|
SATB homeobox 2 |
| chr9_+_48415043 | 0.64 |
ENSDART00000159930
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr21_+_6394929 | 0.63 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr19_-_47323267 | 0.62 |
ENSDART00000190077
|
CU138512.1
|
|
| chr13_-_18122333 | 0.62 |
ENSDART00000128748
|
washc2c
|
WASH complex subunit 2C |
| chr5_+_66433287 | 0.62 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr11_+_23760470 | 0.62 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr19_+_24891747 | 0.62 |
ENSDART00000132209
ENSDART00000193610 |
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr7_-_54679595 | 0.62 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr15_-_18162647 | 0.62 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr17_+_41992054 | 0.61 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr24_-_6078222 | 0.61 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr11_+_42726712 | 0.61 |
ENSDART00000028955
|
tdrd3
|
tudor domain containing 3 |
| chr7_+_19552381 | 0.61 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr6_-_39649504 | 0.61 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr17_+_6625717 | 0.60 |
ENSDART00000190753
ENSDART00000181298 |
BX005321.1
|
|
| chr25_-_25058508 | 0.60 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr11_+_31323746 | 0.59 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr10_+_42733210 | 0.59 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr10_-_43404027 | 0.59 |
ENSDART00000086227
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr5_+_21891305 | 0.59 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr6_-_29305132 | 0.59 |
ENSDART00000132456
|
bivm
|
basic, immunoglobulin-like variable motif containing |
| chr13_+_35339182 | 0.59 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr9_-_47472998 | 0.59 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr22_-_9896180 | 0.58 |
ENSDART00000138343
|
znf990
|
zinc finger protein 990 |
| chr5_-_11809710 | 0.58 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr17_-_2386569 | 0.58 |
ENSDART00000121614
|
PLCB2
|
phospholipase C beta 2 |
| chr5_-_6377865 | 0.58 |
ENSDART00000031775
|
zgc:73226
|
zgc:73226 |
| chr3_+_38540411 | 0.58 |
ENSDART00000154943
|
si:dkey-7f16.3
|
si:dkey-7f16.3 |
| chr6_-_1432200 | 0.57 |
ENSDART00000182901
|
LO018148.1
|
|
| chr14_-_48765262 | 0.57 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr6_-_6487876 | 0.57 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr18_-_30020879 | 0.57 |
ENSDART00000162086
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr15_-_34845414 | 0.57 |
ENSDART00000009892
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr6_-_21189295 | 0.56 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr1_-_46981134 | 0.56 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr23_+_4022620 | 0.56 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr3_+_62205858 | 0.55 |
ENSDART00000126807
|
zgc:173575
|
zgc:173575 |
| chr23_-_25779995 | 0.55 |
ENSDART00000110670
|
si:dkey-21c19.3
|
si:dkey-21c19.3 |
| chr6_+_59832786 | 0.55 |
ENSDART00000154985
ENSDART00000102148 |
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr3_-_10749691 | 0.55 |
ENSDART00000183088
|
CU210890.1
|
|
| chr8_+_24299321 | 0.55 |
ENSDART00000112046
ENSDART00000183558 ENSDART00000183796 ENSDART00000181395 ENSDART00000184555 ENSDART00000172531 ENSDART00000191663 ENSDART00000184103 ENSDART00000183676 |
ZNF335
|
si:ch211-269m15.3 |
| chr3_+_57268363 | 0.54 |
ENSDART00000180725
|
TMEM235 (1 of many)
|
transmembrane protein 235 |
| chr15_+_32297441 | 0.54 |
ENSDART00000153657
|
trim3a
|
tripartite motif containing 3a |
| chr1_-_9195629 | 0.54 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr7_+_66884291 | 0.54 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr22_-_22337382 | 0.53 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr2_+_48073972 | 0.53 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr10_+_15967643 | 0.53 |
ENSDART00000136709
|
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr7_-_11596450 | 0.53 |
ENSDART00000173863
|
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr12_-_48188928 | 0.53 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr24_+_9475809 | 0.53 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr9_-_9348058 | 0.53 |
ENSDART00000132257
|
zgc:113337
|
zgc:113337 |
| chr12_-_9294819 | 0.53 |
ENSDART00000003805
|
pth1rb
|
parathyroid hormone 1 receptor b |
| chr5_-_14373662 | 0.53 |
ENSDART00000183694
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr17_+_16564921 | 0.53 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr3_-_40955780 | 0.52 |
ENSDART00000130130
|
cyp3c3
|
cytochrome P450, family 3, subfamily c, polypeptide 3 |
| chr2_-_17393216 | 0.52 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr10_-_2524297 | 0.52 |
ENSDART00000192475
|
CU856539.1
|
|
| chr22_-_18179214 | 0.52 |
ENSDART00000129576
|
si:ch211-125m10.6
|
si:ch211-125m10.6 |
| chr23_-_26652273 | 0.51 |
ENSDART00000112124
ENSDART00000111029 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr1_-_669717 | 0.51 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr15_-_755023 | 0.51 |
ENSDART00000155594
|
znf1011
|
zinc finger protein 1011 |
Network of associatons between targets according to the STRING database.
First level regulatory network of barx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.4 | 1.5 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.3 | 1.0 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.3 | 1.9 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.2 | 1.0 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.2 | 0.7 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.2 | 2.5 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.2 | 0.7 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.2 | 1.8 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 1.8 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.2 | 0.8 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.2 | 0.8 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 0.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 1.2 | GO:0032475 | otolith formation(GO:0032475) |
| 0.2 | 0.7 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.2 | 1.2 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 | 0.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.4 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 1.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.5 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 0.6 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 1.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.0 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.8 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.4 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 0.7 | GO:1902023 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.5 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.7 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.1 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.5 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.5 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 1.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 1.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.3 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.1 | 0.9 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.1 | 0.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.8 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 0.4 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.1 | 0.9 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 1.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.5 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.6 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.7 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 2.4 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.8 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 2.9 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 1.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.4 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.4 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.3 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.3 | GO:0042138 | mitotic G2 DNA damage checkpoint(GO:0007095) meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.7 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.4 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.6 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.4 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.4 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.4 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.9 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.3 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 1.5 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.5 | GO:0030814 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.3 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.7 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.8 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.7 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.4 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.2 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.7 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 2.3 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.1 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.0 | 0.2 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0008216 | spermidine metabolic process(GO:0008216) polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.4 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.6 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.3 | GO:0033135 | regulation of peptidyl-serine phosphorylation(GO:0033135) |
| 0.0 | 0.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.0 | 0.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.6 | GO:0002685 | regulation of leukocyte migration(GO:0002685) |
| 0.0 | 0.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.3 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.5 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.6 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 0.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 0.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 0.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 2.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.4 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.1 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.6 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.7 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 2.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.3 | 0.9 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.3 | 1.8 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.2 | 0.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 2.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 0.7 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.2 | 0.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 0.8 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.2 | 0.6 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.2 | 1.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.4 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.7 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.9 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.5 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 0.4 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.1 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.5 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 1.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 2.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.7 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.2 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 3.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.4 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 1.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.9 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 5.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 3.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 1.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 3.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0002020 | protease binding(GO:0002020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 1.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.0 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |