Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for barx1
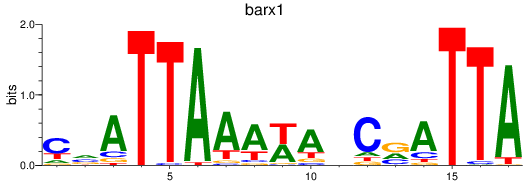
Z-value: 0.84
Transcription factors associated with barx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barx1
|
ENSDARG00000007407 | BARX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barx1 | dr11_v1_chr11_+_27543093_27543093 | -0.17 | 4.8e-01 | Click! |
Activity profile of barx1 motif
Sorted Z-values of barx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_24305877 | 2.56 |
ENSDART00000144226
|
ctsll
|
cathepsin L, like |
| chr12_-_16898140 | 2.18 |
ENSDART00000152656
|
MGC174155
|
Cathepsin L1-like |
| chr12_-_16923162 | 2.10 |
ENSDART00000106072
|
si:dkey-26g8.5
|
si:dkey-26g8.5 |
| chr18_-_49318823 | 1.82 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr12_-_16720432 | 1.82 |
ENSDART00000152261
ENSDART00000152154 |
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr11_-_35171768 | 1.69 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr9_+_21306902 | 1.50 |
ENSDART00000138554
ENSDART00000004108 |
xpo4
|
exportin 4 |
| chr8_+_28358161 | 1.44 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr7_+_32693890 | 1.44 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr2_-_42960353 | 1.41 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr16_-_55028740 | 1.41 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr12_+_27141140 | 1.40 |
ENSDART00000136415
|
hoxb1b
|
homeobox B1b |
| chr4_-_12477224 | 1.37 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr7_+_59169081 | 1.37 |
ENSDART00000167980
|
ostc
|
oligosaccharyltransferase complex subunit |
| chr13_+_7442023 | 1.30 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr9_-_48370645 | 1.19 |
ENSDART00000140185
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr16_+_29509133 | 1.16 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr22_+_26400519 | 1.15 |
ENSDART00000159839
ENSDART00000144585 |
capn8
|
calpain 8 |
| chr3_-_26183699 | 1.12 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr3_+_60721342 | 1.09 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr20_-_26822522 | 1.09 |
ENSDART00000146326
ENSDART00000046764 ENSDART00000103234 ENSDART00000143267 |
gmds
|
GDP-mannose 4,6-dehydratase |
| chr21_+_38855551 | 1.06 |
ENSDART00000171977
|
ddx52
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 |
| chr4_+_19700308 | 1.06 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr12_-_30043809 | 1.05 |
ENSDART00000150220
ENSDART00000075889 ENSDART00000148847 |
trub1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr24_+_36239617 | 1.04 |
ENSDART00000062745
|
riok3
|
RIO kinase 3 (yeast) |
| chr9_-_32753535 | 1.02 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr14_+_989733 | 1.02 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr19_-_11315224 | 1.01 |
ENSDART00000104933
|
eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr5_-_63515210 | 1.01 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr9_-_12401871 | 1.00 |
ENSDART00000191901
|
nup35
|
nucleoporin 35 |
| chr8_+_2487883 | 0.99 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr5_-_41531629 | 0.98 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr9_-_55128839 | 0.96 |
ENSDART00000085135
|
tbl1x
|
transducin beta like 1 X-linked |
| chr12_+_47917971 | 0.96 |
ENSDART00000185933
|
tbata
|
thymus, brain and testes associated |
| chr19_+_16016038 | 0.95 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr5_-_51998708 | 0.94 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr19_-_11950434 | 0.94 |
ENSDART00000024861
|
cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr2_-_9989919 | 0.94 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr22_+_39084829 | 0.91 |
ENSDART00000002826
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr1_+_47459723 | 0.90 |
ENSDART00000015046
|
fstl1a
|
follistatin-like 1a |
| chr11_+_17984354 | 0.89 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr25_+_14507567 | 0.89 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr7_+_72003301 | 0.89 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr22_-_26005894 | 0.89 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr16_-_48430272 | 0.88 |
ENSDART00000005927
|
rad21a
|
RAD21 cohesin complex component a |
| chr13_-_36535128 | 0.88 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr11_-_3987885 | 0.87 |
ENSDART00000058735
|
glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr16_+_40576679 | 0.87 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr22_+_10651726 | 0.86 |
ENSDART00000145459
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr7_-_30174882 | 0.86 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr4_+_15944245 | 0.86 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr10_-_29827000 | 0.86 |
ENSDART00000131418
|
zpr1
|
ZPR1 zinc finger |
| chr22_-_10641873 | 0.85 |
ENSDART00000064772
|
cyb561d2
|
cytochrome b561 family, member D2 |
| chr2_-_57837838 | 0.85 |
ENSDART00000010699
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr10_-_43294933 | 0.84 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr6_+_25257728 | 0.84 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr2_+_9990491 | 0.84 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr22_+_26443235 | 0.84 |
ENSDART00000044085
|
zgc:92480
|
zgc:92480 |
| chr21_-_25604603 | 0.84 |
ENSDART00000133134
ENSDART00000139460 |
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr3_-_26805455 | 0.84 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr1_-_11104805 | 0.83 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr3_-_26806032 | 0.83 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr10_-_3138403 | 0.83 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr16_+_14812585 | 0.81 |
ENSDART00000134087
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr4_+_42556555 | 0.81 |
ENSDART00000168536
|
znf1053
|
zinc finger protein 1053 |
| chr17_+_8799451 | 0.80 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr19_+_10847118 | 0.79 |
ENSDART00000189784
|
apoa4a
|
apolipoprotein A-IV a |
| chr3_-_3428938 | 0.79 |
ENSDART00000179811
ENSDART00000115282 ENSDART00000192263 ENSDART00000046454 |
A2ML1 (1 of many)
|
zgc:171445 |
| chr19_+_30884706 | 0.78 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr1_-_21723329 | 0.78 |
ENSDART00000137138
|
si:ch211-134c9.2
|
si:ch211-134c9.2 |
| chr1_+_10318089 | 0.78 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr22_+_19640309 | 0.77 |
ENSDART00000061725
ENSDART00000140819 |
rgmd
|
RGM domain family, member D |
| chr5_+_12528693 | 0.77 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr3_-_26184018 | 0.77 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr4_-_72296520 | 0.76 |
ENSDART00000182638
|
si:cabz01071911.3
|
si:cabz01071911.3 |
| chr22_-_15587360 | 0.76 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr5_+_72137331 | 0.76 |
ENSDART00000149195
|
exosc2
|
exosome component 2 |
| chr13_+_18321140 | 0.75 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr10_+_9410304 | 0.75 |
ENSDART00000080843
|
ndufa8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
| chr24_-_20926812 | 0.75 |
ENSDART00000130958
|
ccdc58
|
coiled-coil domain containing 58 |
| chr8_-_2230128 | 0.75 |
ENSDART00000140427
|
si:dkeyp-117b11.2
|
si:dkeyp-117b11.2 |
| chr19_+_42609132 | 0.75 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr11_+_17984167 | 0.74 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr17_-_20228610 | 0.74 |
ENSDART00000125758
|
ebf3b
|
early B cell factor 3b |
| chr5_-_22602780 | 0.73 |
ENSDART00000011699
|
nono
|
non-POU domain containing, octamer-binding |
| chr19_-_868187 | 0.73 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr15_+_21254800 | 0.73 |
ENSDART00000142902
|
usf1
|
upstream transcription factor 1 |
| chr17_-_29224908 | 0.73 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr12_+_11007286 | 0.73 |
ENSDART00000166796
|
raraa
|
retinoic acid receptor, alpha a |
| chr2_-_32237916 | 0.73 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr1_+_9860381 | 0.72 |
ENSDART00000054848
|
pmm2
|
phosphomannomutase 2 |
| chr8_+_2487250 | 0.72 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr20_+_53441935 | 0.71 |
ENSDART00000175214
|
apobb.2
|
apolipoprotein Bb, tandem duplicate 2 |
| chr1_+_14020445 | 0.71 |
ENSDART00000079716
|
hpf1
|
histone PARylation factor 1 |
| chr7_+_15308219 | 0.71 |
ENSDART00000165683
|
mespba
|
mesoderm posterior ba |
| chr25_-_18435481 | 0.70 |
ENSDART00000004771
|
poc1b
|
POC1 centriolar protein B |
| chr8_-_8489886 | 0.70 |
ENSDART00000183334
|
abt1
|
activator of basal transcription 1 |
| chr18_+_36582815 | 0.70 |
ENSDART00000059311
|
siae
|
sialic acid acetylesterase |
| chr12_+_47663419 | 0.70 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr24_-_21090447 | 0.70 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr9_+_4675193 | 0.70 |
ENSDART00000081326
|
prpf40a
|
PRP40 pre-mRNA processing factor 40 homolog A |
| chr1_+_25696798 | 0.69 |
ENSDART00000054228
|
lrata
|
lecithin retinol acyltransferase a |
| chr1_-_53468160 | 0.69 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr11_-_36263886 | 0.69 |
ENSDART00000140397
|
nfya
|
nuclear transcription factor Y, alpha |
| chr6_-_15757867 | 0.69 |
ENSDART00000063665
|
ackr3b
|
atypical chemokine receptor 3b |
| chr16_-_7379328 | 0.69 |
ENSDART00000060447
|
med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr3_+_15893039 | 0.69 |
ENSDART00000055780
|
jpt2
|
Jupiter microtubule associated homolog 2 |
| chr16_+_14201401 | 0.69 |
ENSDART00000113679
|
dap3
|
death associated protein 3 |
| chr24_+_18299506 | 0.69 |
ENSDART00000172056
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr19_+_31585341 | 0.68 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr9_-_41040492 | 0.68 |
ENSDART00000163164
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr16_-_25608453 | 0.68 |
ENSDART00000140140
|
zgc:110410
|
zgc:110410 |
| chr21_-_44104600 | 0.68 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr16_-_35952789 | 0.66 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr24_+_35387517 | 0.66 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr6_-_11523987 | 0.66 |
ENSDART00000189363
|
gulp1b
|
GULP, engulfment adaptor PTB domain containing 1b |
| chr2_-_26469065 | 0.66 |
ENSDART00000099247
ENSDART00000099248 |
rabggtb
|
Rab geranylgeranyltransferase, beta subunit |
| chr6_-_40768654 | 0.66 |
ENSDART00000184668
ENSDART00000146470 |
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr5_-_54554583 | 0.66 |
ENSDART00000158865
ENSDART00000158069 |
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr4_-_52165969 | 0.66 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr3_+_40289418 | 0.65 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr5_+_42280372 | 0.65 |
ENSDART00000142855
|
tbx6l
|
T-box 6, like |
| chr4_+_1761029 | 0.65 |
ENSDART00000026963
|
pmpcb
|
peptidase (mitochondrial processing) beta |
| chr21_-_13231101 | 0.65 |
ENSDART00000190943
|
setb
|
SET nuclear proto-oncogene b |
| chr9_-_18911608 | 0.65 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr17_-_5426458 | 0.64 |
ENSDART00000157937
|
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr13_+_18311410 | 0.64 |
ENSDART00000036718
ENSDART00000132073 |
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr23_+_36460239 | 0.64 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr8_-_45416187 | 0.64 |
ENSDART00000148676
|
mak16
|
MAK16 homolog (S. cerevisiae) |
| chr17_+_8799661 | 0.64 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr16_+_33938227 | 0.63 |
ENSDART00000166254
|
gpn2
|
GPN-loop GTPase 2 |
| chr16_-_27566552 | 0.63 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr18_+_3572314 | 0.63 |
ENSDART00000169814
ENSDART00000157819 |
serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr2_+_27386617 | 0.63 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr6_-_59505589 | 0.63 |
ENSDART00000170685
|
gli1
|
GLI family zinc finger 1 |
| chr11_+_6136220 | 0.63 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr21_-_26071773 | 0.63 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr19_-_7441948 | 0.62 |
ENSDART00000003544
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr9_+_44994214 | 0.62 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr3_+_32410746 | 0.62 |
ENSDART00000025496
|
rras
|
RAS related |
| chr25_+_19095231 | 0.62 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr3_-_12930217 | 0.61 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr21_-_1635268 | 0.61 |
ENSDART00000151258
|
zgc:152948
|
zgc:152948 |
| chr1_+_513986 | 0.61 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr18_-_34549721 | 0.61 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr13_+_5978809 | 0.61 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr4_-_4250317 | 0.61 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr11_-_16152105 | 0.61 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr13_-_50341004 | 0.60 |
ENSDART00000038120
|
cacul1
|
CDK2 associated cullin domain 1 |
| chr21_-_21020708 | 0.60 |
ENSDART00000064032
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr3_+_28502419 | 0.60 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr21_-_26071142 | 0.60 |
ENSDART00000004740
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr17_+_4030493 | 0.59 |
ENSDART00000151849
|
hao1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr7_+_20471315 | 0.59 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr2_-_29994726 | 0.59 |
ENSDART00000163350
|
cnpy1
|
canopy1 |
| chr14_-_7141550 | 0.59 |
ENSDART00000172020
|
trpt1
|
tRNA phosphotransferase 1 |
| chr17_+_45639247 | 0.58 |
ENSDART00000153669
|
vcpkmt
|
valosin containing protein lysine (K) methyltransferase |
| chr1_-_28749604 | 0.58 |
ENSDART00000148522
|
GK3P
|
zgc:172295 |
| chr25_+_186583 | 0.58 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr1_-_45584407 | 0.57 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr16_-_10223741 | 0.57 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr1_+_26676758 | 0.57 |
ENSDART00000152299
|
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr1_-_35924495 | 0.57 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr22_-_24818066 | 0.57 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr13_+_35637048 | 0.57 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr17_-_7792376 | 0.56 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr9_-_40683722 | 0.56 |
ENSDART00000141979
ENSDART00000181228 |
bard1
|
BRCA1 associated RING domain 1 |
| chr25_+_35019693 | 0.56 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr20_-_43723860 | 0.56 |
ENSDART00000122051
|
mixl1
|
Mix paired-like homeobox |
| chr6_+_33931095 | 0.56 |
ENSDART00000191909
|
orc1
|
origin recognition complex, subunit 1 |
| chr25_+_2263857 | 0.56 |
ENSDART00000076439
|
yars2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr6_-_58764672 | 0.56 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr1_-_45215343 | 0.56 |
ENSDART00000014727
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr17_+_25849332 | 0.56 |
ENSDART00000191994
|
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr14_-_35462148 | 0.56 |
ENSDART00000045809
|
srpx2
|
sushi-repeat containing protein, X-linked 2 |
| chr23_+_44080382 | 0.56 |
ENSDART00000011715
|
zgc:56304
|
zgc:56304 |
| chr10_+_8968203 | 0.56 |
ENSDART00000110443
ENSDART00000080772 |
fstb
|
follistatin b |
| chr19_-_23918136 | 0.56 |
ENSDART00000079842
|
chtopa
|
chromatin target of PRMT1a |
| chr21_+_34122801 | 0.55 |
ENSDART00000182627
|
hmgb3b
|
high mobility group box 3b |
| chr13_+_3938526 | 0.55 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr21_+_5192016 | 0.55 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr15_-_47865063 | 0.55 |
ENSDART00000151600
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr16_+_16968682 | 0.55 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr8_-_18537866 | 0.55 |
ENSDART00000148802
ENSDART00000148962 ENSDART00000149506 |
nexn
|
nexilin (F actin binding protein) |
| chr6_+_23809163 | 0.54 |
ENSDART00000170402
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr19_+_30884960 | 0.54 |
ENSDART00000140603
ENSDART00000183224 ENSDART00000135484 ENSDART00000139599 |
yars
|
tyrosyl-tRNA synthetase |
| chr21_-_929293 | 0.54 |
ENSDART00000006419
|
txnl1
|
thioredoxin-like 1 |
| chr16_+_17715243 | 0.54 |
ENSDART00000149437
ENSDART00000149596 |
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr1_-_8652648 | 0.54 |
ENSDART00000138324
ENSDART00000141407 ENSDART00000054987 |
actb1
|
actin, beta 1 |
| chr2_-_39015130 | 0.54 |
ENSDART00000044331
|
copb2
|
coatomer protein complex, subunit beta 2 |
| chr17_+_13088800 | 0.54 |
ENSDART00000149779
|
gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr15_+_2857556 | 0.54 |
ENSDART00000157758
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr21_-_929448 | 0.54 |
ENSDART00000133976
|
txnl1
|
thioredoxin-like 1 |
| chr16_+_35870456 | 0.54 |
ENSDART00000184321
|
thrap3a
|
thyroid hormone receptor associated protein 3a |
| chr1_-_33557915 | 0.54 |
ENSDART00000075632
|
creb1a
|
cAMP responsive element binding protein 1a |
| chr7_+_32898224 | 0.53 |
ENSDART00000132020
|
tssc4
|
tumor suppressing subtransferable candidate 4 |
| chr19_+_20778011 | 0.53 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr3_-_61362398 | 0.53 |
ENSDART00000156177
|
si:dkey-111k8.3
|
si:dkey-111k8.3 |
| chr11_-_41853874 | 0.53 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr3_-_61387273 | 0.53 |
ENSDART00000156479
|
znf1143
|
zinc finger protein 1143 |
Network of associatons between targets according to the STRING database.
First level regulatory network of barx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.3 | 1.0 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.3 | 1.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 1.0 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.3 | 1.7 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 1.6 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.3 | 1.6 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.3 | 1.3 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.3 | 0.8 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.3 | 1.5 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 0.7 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.2 | 1.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 0.7 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.2 | 1.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 0.6 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 1.0 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 0.6 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 1.4 | GO:0021570 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.2 | 0.6 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.2 | 2.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 0.9 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.2 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 0.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 0.7 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.2 | 0.5 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.2 | 1.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 0.5 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 1.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 0.5 | GO:0001113 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.7 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 1.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.9 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 1.6 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.5 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.5 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.3 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.1 | 1.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.4 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 1.4 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.8 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 1.0 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.3 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.6 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 0.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.5 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 2.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.3 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.1 | 0.8 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.4 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.8 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.4 | GO:0051591 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.0 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.6 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.9 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 0.6 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.4 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.1 | 0.5 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.1 | 0.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.4 | GO:0032447 | protein urmylation(GO:0032447) tRNA thio-modification(GO:0034227) |
| 0.1 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 1.0 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 0.3 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.3 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 1.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 1.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 1.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.3 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) negative regulation of retinoic acid receptor signaling pathway(GO:0048387) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 1.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.4 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.6 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.6 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.1 | 0.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.3 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.3 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 0.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.2 | GO:0071295 | cellular response to nutrient(GO:0031670) cellular response to vitamin(GO:0071295) cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 2.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 1.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.5 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.4 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.1 | GO:0030237 | female sex determination(GO:0030237) |
| 0.0 | 0.2 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 1.1 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.3 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.3 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.5 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0032206 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.3 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.9 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 1.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.4 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.7 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 1.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.0 | 0.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.7 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.5 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.5 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.3 | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902253) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.7 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.5 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 1.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.4 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.5 | GO:0007379 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.4 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 2.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.0 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.1 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.2 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.1 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.6 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 1.1 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.7 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.9 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 1.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.3 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.9 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.5 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 1.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 1.4 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.2 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.9 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.9 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.0 | 1.3 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.4 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.3 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.0 | 0.4 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.3 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:1902806 | regulation of cell cycle G1/S phase transition(GO:1902806) regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.8 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.0 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.0 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.0 | 0.1 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 0.6 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.9 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.6 | GO:0009566 | single fertilization(GO:0007338) fertilization(GO:0009566) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.9 | GO:0048884 | neuromast development(GO:0048884) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0031647 | regulation of protein stability(GO:0031647) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.3 | 0.9 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.3 | 0.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 1.0 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 0.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 1.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 1.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 0.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.6 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.6 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.7 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.1 | 0.5 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.9 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.4 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.4 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 1.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 1.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.3 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.7 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.5 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.6 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.7 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.3 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 1.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.2 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.6 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 4.1 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 2.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 1.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 0.7 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.2 | 0.7 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.2 | 0.9 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.2 | 0.7 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.2 | 1.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 1.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 1.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 1.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 0.7 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 2.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 1.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.6 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.5 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 0.9 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.1 | 1.1 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.5 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 2.0 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.5 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 0.8 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.3 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.5 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.3 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.3 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 0.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.5 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.4 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.7 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 1.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.5 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.4 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.8 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.2 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.3 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 4.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 1.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 1.0 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 1.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 1.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.0 | 4.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.8 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.9 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.5 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.7 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 1.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.0 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.2 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 23.7 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.6 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 1.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.9 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |