Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
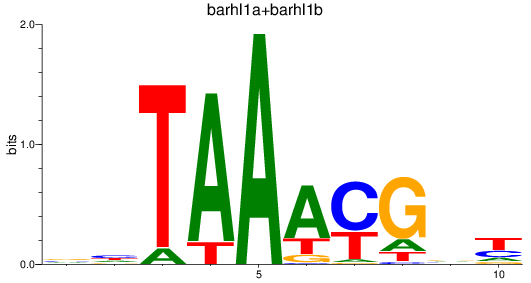
Results for barhl1a+barhl1b
Z-value: 2.71
Transcription factors associated with barhl1a+barhl1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barhl1b
|
ENSDARG00000019013 | BarH-like homeobox 1b |
|
barhl1a
|
ENSDARG00000035508 | BarH-like homeobox 1a |
|
barhl1a
|
ENSDARG00000110061 | BarH-like homeobox 1a |
|
barhl1a
|
ENSDARG00000112355 | BarH-like homeobox 1a |
|
barhl1b
|
ENSDARG00000113145 | BarH-like homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barhl1b | dr11_v1_chr21_-_17482465_17482465 | -0.40 | 9.0e-02 | Click! |
| barhl1a | dr11_v1_chr5_-_29750377_29750377 | -0.06 | 8.1e-01 | Click! |
Activity profile of barhl1a+barhl1b motif
Sorted Z-values of barhl1a+barhl1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_8362419 | 15.66 |
ENSDART00000142752
ENSDART00000135810 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr22_+_14093306 | 12.39 |
ENSDART00000080329
ENSDART00000115383 |
he1.1
|
hatching enzyme 1, tandem duplicate 1 |
| chr3_-_19368435 | 8.61 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr4_+_15006217 | 7.00 |
ENSDART00000090837
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr19_+_15444210 | 5.56 |
ENSDART00000142509
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr16_-_32303835 | 5.55 |
ENSDART00000191408
|
mms22l
|
MMS22-like, DNA repair protein |
| chr7_+_34236238 | 5.45 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr4_+_73606482 | 5.27 |
ENSDART00000150765
|
si:ch211-165i18.2
|
si:ch211-165i18.2 |
| chr3_+_31093455 | 5.22 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr19_-_47571456 | 5.22 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr7_-_54678289 | 5.03 |
ENSDART00000170774
|
ccnd1
|
cyclin D1 |
| chr3_-_61387273 | 4.99 |
ENSDART00000156479
|
znf1143
|
zinc finger protein 1143 |
| chr10_-_35257458 | 4.99 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr8_+_30452945 | 4.91 |
ENSDART00000062303
|
foxd5
|
forkhead box D5 |
| chr24_-_41797681 | 4.84 |
ENSDART00000169643
|
arhgap28
|
Rho GTPase activating protein 28 |
| chr23_+_31913292 | 4.72 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr20_-_53992952 | 4.72 |
ENSDART00000170138
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr20_+_33294428 | 4.65 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr2_-_10098191 | 4.62 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr16_+_40576679 | 4.48 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr5_-_69437422 | 4.43 |
ENSDART00000073676
|
isca1
|
iron-sulfur cluster assembly 1 |
| chr4_-_12477224 | 4.39 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr13_-_35908275 | 4.30 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr20_-_32148901 | 4.28 |
ENSDART00000153405
ENSDART00000048537 ENSDART00000152984 |
cep57l1
|
centrosomal protein 57, like 1 |
| chr6_+_3473657 | 4.28 |
ENSDART00000011785
|
rad54l
|
RAD54 like |
| chr13_+_15701849 | 4.25 |
ENSDART00000003517
|
trmt61a
|
tRNA methyltransferase 61A |
| chr23_-_25779995 | 4.21 |
ENSDART00000110670
|
si:dkey-21c19.3
|
si:dkey-21c19.3 |
| chr5_+_24087035 | 4.10 |
ENSDART00000183644
|
tp53
|
tumor protein p53 |
| chr14_-_17072736 | 4.08 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr20_-_38827623 | 4.08 |
ENSDART00000153310
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr23_-_10722664 | 4.07 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr2_+_27386617 | 4.05 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr12_+_14149686 | 4.04 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr16_+_29376751 | 4.04 |
ENSDART00000168856
ENSDART00000162502 ENSDART00000050535 |
rrnad1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr1_+_49352900 | 4.00 |
ENSDART00000008468
|
msx1b
|
muscle segment homeobox 1b |
| chr23_+_35713557 | 3.99 |
ENSDART00000123518
|
tuba1c
|
tubulin, alpha 1c |
| chr22_-_21314821 | 3.99 |
ENSDART00000105546
ENSDART00000135388 |
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr2_-_38080075 | 3.96 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr22_-_10541372 | 3.92 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr5_-_32505109 | 3.85 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr5_+_32831561 | 3.83 |
ENSDART00000169358
ENSDART00000192078 |
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr3_+_22442445 | 3.83 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr3_-_50066499 | 3.81 |
ENSDART00000056618
ENSDART00000154561 |
mrpl12
|
mitochondrial ribosomal protein L12 |
| chr16_-_31824525 | 3.79 |
ENSDART00000058737
|
cdc42l
|
cell division cycle 42, like |
| chr5_-_32505276 | 3.76 |
ENSDART00000034705
ENSDART00000187597 |
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr1_-_31515746 | 3.75 |
ENSDART00000190886
|
cenpk
|
centromere protein K |
| chr18_+_29898955 | 3.74 |
ENSDART00000064080
|
cenpn
|
centromere protein N |
| chr19_+_3213914 | 3.72 |
ENSDART00000193144
|
zgc:86598
|
zgc:86598 |
| chr6_+_11760749 | 3.71 |
ENSDART00000112212
|
zswim2
|
zinc finger, SWIM-type containing 2 |
| chr19_+_41509956 | 3.68 |
ENSDART00000171318
ENSDART00000172311 |
si:ch211-57n23.1
|
si:ch211-57n23.1 |
| chr7_+_25000060 | 3.63 |
ENSDART00000039265
ENSDART00000141814 |
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr2_-_29994726 | 3.59 |
ENSDART00000163350
|
cnpy1
|
canopy1 |
| chr16_+_48626780 | 3.58 |
ENSDART00000157364
|
pbx2
|
pre-B-cell leukemia homeobox 2 |
| chr2_-_17115256 | 3.47 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr11_-_40519886 | 3.45 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr1_-_55166511 | 3.44 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr2_-_16449504 | 3.41 |
ENSDART00000144801
|
atr
|
ATR serine/threonine kinase |
| chr13_-_31296358 | 3.41 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr23_+_24955394 | 3.31 |
ENSDART00000142124
|
nol9
|
nucleolar protein 9 |
| chr16_-_34195002 | 3.21 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr18_+_7639401 | 3.19 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr12_+_23664335 | 3.17 |
ENSDART00000111334
|
mtpap
|
mitochondrial poly(A) polymerase |
| chr4_-_9350585 | 3.15 |
ENSDART00000148155
|
si:ch211-125a15.1
|
si:ch211-125a15.1 |
| chr5_+_60847823 | 3.14 |
ENSDART00000074426
|
lig3
|
ligase III, DNA, ATP-dependent |
| chr2_+_32826235 | 3.12 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr8_+_20863025 | 3.12 |
ENSDART00000015891
ENSDART00000187089 ENSDART00000146989 |
fzr1a
|
fizzy/cell division cycle 20 related 1a |
| chr2_-_26469065 | 3.12 |
ENSDART00000099247
ENSDART00000099248 |
rabggtb
|
Rab geranylgeranyltransferase, beta subunit |
| chr9_+_38314466 | 3.09 |
ENSDART00000048753
|
ddx18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr14_+_21820034 | 3.08 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr19_-_11315224 | 3.02 |
ENSDART00000104933
|
eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr20_-_54924593 | 2.97 |
ENSDART00000151522
|
si:dkey-15f23.1
|
si:dkey-15f23.1 |
| chr9_-_34300707 | 2.95 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr7_+_25126629 | 2.92 |
ENSDART00000077217
|
zgc:101765
|
zgc:101765 |
| chr13_+_6189203 | 2.92 |
ENSDART00000109665
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr13_+_6188759 | 2.87 |
ENSDART00000161062
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr1_-_55116453 | 2.86 |
ENSDART00000142348
|
sertad2a
|
SERTA domain containing 2a |
| chr19_+_29808471 | 2.86 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr16_+_26732086 | 2.85 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr3_-_15352303 | 2.84 |
ENSDART00000104338
ENSDART00000145919 ENSDART00000132135 |
pitpnbl
|
phosphatidylinositol transfer protein, beta, like |
| chr22_-_38258053 | 2.83 |
ENSDART00000132516
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr11_+_30295582 | 2.80 |
ENSDART00000122424
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr23_+_31912882 | 2.77 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr17_-_45387134 | 2.76 |
ENSDART00000010975
|
tmem206
|
transmembrane protein 206 |
| chr19_-_33274978 | 2.75 |
ENSDART00000020301
ENSDART00000114714 |
fam92a1
|
family with sequence similarity 92, member A1 |
| chr18_+_5308392 | 2.75 |
ENSDART00000179072
|
DUT
|
deoxyuridine triphosphatase |
| chr7_-_20464133 | 2.74 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr12_-_20409794 | 2.66 |
ENSDART00000077936
|
lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr13_-_35907768 | 2.64 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr13_+_16279890 | 2.64 |
ENSDART00000101775
ENSDART00000057948 |
anxa11a
|
annexin A11a |
| chr6_-_6976096 | 2.63 |
ENSDART00000151822
ENSDART00000039443 ENSDART00000177960 |
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr3_+_54761569 | 2.63 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr9_+_52411530 | 2.62 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr22_+_17120473 | 2.61 |
ENSDART00000170076
ENSDART00000090217 |
frem1b
|
Fras1 related extracellular matrix 1b |
| chr5_+_36900157 | 2.60 |
ENSDART00000183533
ENSDART00000051184 |
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr14_+_21686207 | 2.59 |
ENSDART00000034438
|
ran
|
RAN, member RAS oncogene family |
| chr25_+_1732838 | 2.59 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr23_+_32011768 | 2.58 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr23_+_19813677 | 2.58 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr3_+_17910569 | 2.57 |
ENSDART00000080946
|
ttc25
|
tetratricopeptide repeat domain 25 |
| chr13_+_9896368 | 2.57 |
ENSDART00000137388
|
si:ch211-117n7.8
|
si:ch211-117n7.8 |
| chr16_+_20055878 | 2.57 |
ENSDART00000146436
|
ankrd28b
|
ankyrin repeat domain 28b |
| chr13_+_9896845 | 2.56 |
ENSDART00000169076
|
si:ch211-117n7.8
|
si:ch211-117n7.8 |
| chr16_-_43344859 | 2.55 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr3_+_12554801 | 2.53 |
ENSDART00000167177
|
ccnf
|
cyclin F |
| chr11_+_6136220 | 2.53 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr18_+_6536598 | 2.52 |
ENSDART00000149350
|
fkbp4
|
FK506 binding protein 4 |
| chr10_+_10728870 | 2.52 |
ENSDART00000109282
|
swi5
|
SWI5 homologous recombination repair protein |
| chr14_-_38929885 | 2.51 |
ENSDART00000148737
|
btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr19_-_20777351 | 2.50 |
ENSDART00000019206
|
ngly1
|
N-glycanase 1 |
| chr25_-_26753196 | 2.49 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr23_-_31913231 | 2.48 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr2_+_22495274 | 2.48 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 VRAC subunit Da |
| chr16_+_15114645 | 2.48 |
ENSDART00000158483
|
mtbp
|
MDM2 binding protein |
| chr20_-_20901540 | 2.47 |
ENSDART00000010564
ENSDART00000131273 |
xrcc3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr21_-_25565392 | 2.47 |
ENSDART00000144917
ENSDART00000180102 |
si:dkey-17e16.10
|
si:dkey-17e16.10 |
| chr15_-_14193926 | 2.46 |
ENSDART00000162707
|
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr17_-_33716688 | 2.45 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr12_+_16087077 | 2.44 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr1_-_31516091 | 2.44 |
ENSDART00000139828
ENSDART00000146567 |
cenpk
|
centromere protein K |
| chr7_-_24644893 | 2.43 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr2_+_16449627 | 2.43 |
ENSDART00000005381
|
zgc:110269
|
zgc:110269 |
| chr11_-_33618612 | 2.42 |
ENSDART00000033980
|
lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr16_-_35415004 | 2.40 |
ENSDART00000170401
ENSDART00000157832 ENSDART00000170048 |
taf12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr9_-_33477588 | 2.39 |
ENSDART00000144150
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr10_-_13116337 | 2.39 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr18_-_7448047 | 2.38 |
ENSDART00000193213
ENSDART00000131940 ENSDART00000186944 ENSDART00000052803 |
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr12_+_28910762 | 2.36 |
ENSDART00000076342
ENSDART00000160939 ENSDART00000076572 |
rnf40
|
ring finger protein 40 |
| chr7_-_24112484 | 2.36 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr25_+_17590095 | 2.35 |
ENSDART00000009767
|
vac14
|
vac14 homolog (S. cerevisiae) |
| chr7_+_56517402 | 2.35 |
ENSDART00000073597
|
dhodh
|
dihydroorotate dehydrogenase |
| chr2_-_37103622 | 2.35 |
ENSDART00000137849
|
zgc:101744
|
zgc:101744 |
| chr3_+_7040363 | 2.35 |
ENSDART00000157805
|
BX000701.2
|
|
| chr7_+_38260434 | 2.35 |
ENSDART00000052351
|
cnep1r1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr3_-_32859335 | 2.35 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr19_-_874888 | 2.33 |
ENSDART00000007206
|
eomesa
|
eomesodermin homolog a |
| chr9_+_500052 | 2.31 |
ENSDART00000166707
|
CU984600.1
|
|
| chr15_-_41689684 | 2.31 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr3_+_22035863 | 2.30 |
ENSDART00000177169
|
cdc27
|
cell division cycle 27 |
| chr25_-_12923482 | 2.28 |
ENSDART00000161754
|
CR450808.1
|
|
| chr21_+_10756154 | 2.27 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr23_-_20345473 | 2.27 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr17_+_24446353 | 2.27 |
ENSDART00000140467
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr23_-_24955135 | 2.24 |
ENSDART00000136347
ENSDART00000144903 |
zbtb48
|
zinc finger and BTB domain containing 48 |
| chr13_-_35808904 | 2.24 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr5_+_51833132 | 2.21 |
ENSDART00000167491
|
papd4
|
PAP associated domain containing 4 |
| chr18_-_34549721 | 2.21 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr11_-_25461336 | 2.21 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr11_+_37612214 | 2.21 |
ENSDART00000172899
ENSDART00000077496 |
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr22_+_10010292 | 2.21 |
ENSDART00000180096
|
BX324216.4
|
|
| chr10_-_11397590 | 2.21 |
ENSDART00000064212
|
srek1ip1
|
SREK1-interacting protein 1 |
| chr3_+_22036113 | 2.19 |
ENSDART00000132190
|
cdc27
|
cell division cycle 27 |
| chr1_+_29267841 | 2.17 |
ENSDART00000085648
|
lig4
|
ligase IV, DNA, ATP-dependent |
| chr3_+_22443313 | 2.17 |
ENSDART00000156450
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr12_+_34119439 | 2.17 |
ENSDART00000032821
|
cyth1b
|
cytohesin 1b |
| chr16_+_20056030 | 2.16 |
ENSDART00000027020
|
ankrd28b
|
ankyrin repeat domain 28b |
| chr5_-_33280699 | 2.16 |
ENSDART00000183838
|
kyat1
|
kynurenine aminotransferase 1 |
| chr5_+_57714902 | 2.15 |
ENSDART00000182860
|
ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr5_-_56943064 | 2.14 |
ENSDART00000146991
|
si:ch211-127d4.3
|
si:ch211-127d4.3 |
| chr5_+_53580846 | 2.14 |
ENSDART00000184967
ENSDART00000161751 |
taf6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr8_-_25761544 | 2.13 |
ENSDART00000078152
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr25_-_3347418 | 2.12 |
ENSDART00000082385
|
golt1bb
|
golgi transport 1Bb |
| chr3_+_45365098 | 2.11 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr16_+_11779534 | 2.10 |
ENSDART00000133497
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr5_+_32141790 | 2.08 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr4_-_18840487 | 2.07 |
ENSDART00000066978
|
cbll1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr9_+_8968702 | 2.07 |
ENSDART00000008490
|
ube2al
|
ubiquitin conjugating enzyme E2 A, like |
| chr18_+_48423973 | 2.07 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr10_+_25947946 | 2.06 |
ENSDART00000064393
|
ufm1
|
ubiquitin-fold modifier 1 |
| chr19_+_46237665 | 2.05 |
ENSDART00000159391
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr20_+_26556372 | 2.05 |
ENSDART00000187179
ENSDART00000157291 |
irf4b
|
interferon regulatory factor 4b |
| chr9_-_21488976 | 2.05 |
ENSDART00000080404
|
mphosph8
|
M-phase phosphoprotein 8 |
| chr10_+_42733210 | 2.04 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr6_+_296130 | 2.04 |
ENSDART00000073985
|
rbfox2
|
RNA binding fox-1 homolog 2 |
| chr14_+_7892383 | 2.03 |
ENSDART00000063837
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr23_+_20431388 | 2.03 |
ENSDART00000132920
ENSDART00000102963 ENSDART00000109899 ENSDART00000140219 |
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr2_-_29923403 | 2.03 |
ENSDART00000144672
|
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr17_-_45386823 | 2.02 |
ENSDART00000156002
|
tmem206
|
transmembrane protein 206 |
| chr17_+_24597001 | 2.01 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr20_+_26556174 | 2.01 |
ENSDART00000138492
|
irf4b
|
interferon regulatory factor 4b |
| chr19_+_24891747 | 2.01 |
ENSDART00000132209
ENSDART00000193610 |
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr2_+_6243144 | 2.00 |
ENSDART00000058258
|
gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr12_+_20583552 | 2.00 |
ENSDART00000170035
|
arsg
|
arylsulfatase G |
| chr13_+_22717939 | 1.99 |
ENSDART00000188288
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr3_-_32590164 | 1.99 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr21_+_37090795 | 1.99 |
ENSDART00000085786
|
znf346
|
zinc finger protein 346 |
| chr24_+_16905188 | 1.98 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr2_+_307665 | 1.96 |
ENSDART00000082083
|
zgc:113452
|
zgc:113452 |
| chr15_+_34699585 | 1.96 |
ENSDART00000017569
|
tnfb
|
tumor necrosis factor b (TNF superfamily, member 2) |
| chr19_+_2685779 | 1.96 |
ENSDART00000160533
ENSDART00000097531 |
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr6_-_52348562 | 1.95 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr20_-_10120442 | 1.95 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr21_-_36396334 | 1.94 |
ENSDART00000183627
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr5_-_32890807 | 1.93 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr18_+_49248389 | 1.93 |
ENSDART00000059285
ENSDART00000142004 ENSDART00000132751 |
yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr7_+_26466826 | 1.92 |
ENSDART00000058908
|
mpdu1b
|
mannose-P-dolichol utilization defect 1b |
| chr10_-_8032885 | 1.92 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr10_+_10386435 | 1.92 |
ENSDART00000179214
ENSDART00000189799 ENSDART00000193875 |
sardh
|
sarcosine dehydrogenase |
| chr23_+_27051919 | 1.91 |
ENSDART00000054237
|
ptges3a
|
prostaglandin E synthase 3a (cytosolic) |
| chr17_+_24446533 | 1.90 |
ENSDART00000131417
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr2_-_32386791 | 1.87 |
ENSDART00000056634
|
ubtfl
|
upstream binding transcription factor, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of barhl1a+barhl1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 1.4 | 15.7 | GO:0045453 | bone resorption(GO:0045453) |
| 1.4 | 4.1 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 1.4 | 4.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 1.3 | 5.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 1.2 | 6.0 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 1.2 | 4.7 | GO:0045428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 1.2 | 5.8 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 1.1 | 5.5 | GO:0043111 | replication fork arrest(GO:0043111) |
| 1.1 | 5.3 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 1.0 | 4.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 1.0 | 5.0 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.9 | 3.8 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.8 | 2.5 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.8 | 4.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.8 | 2.4 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.8 | 2.4 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.8 | 2.3 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.8 | 2.3 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.7 | 6.8 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.7 | 2.0 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.6 | 2.6 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.6 | 1.9 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.6 | 1.9 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.6 | 4.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.6 | 9.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.6 | 5.4 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.6 | 3.0 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.6 | 1.8 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.5 | 3.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.5 | 4.1 | GO:0032196 | transposition(GO:0032196) |
| 0.5 | 3.0 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.5 | 1.9 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.5 | 3.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.5 | 5.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.5 | 2.3 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.5 | 5.6 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.5 | 3.2 | GO:1901673 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.4 | 3.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.4 | 5.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.4 | 2.9 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.4 | 3.5 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.4 | 1.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.4 | 1.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.4 | 1.1 | GO:0060306 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.4 | 5.0 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.3 | 5.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.3 | 2.4 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.3 | 1.0 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.3 | 2.5 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.3 | 4.9 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.3 | 2.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.3 | 2.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.3 | 1.2 | GO:0021795 | substrate-dependent cell migration(GO:0006929) pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.3 | 1.4 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.3 | 2.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 2.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.3 | 2.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.3 | 1.9 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 1.8 | GO:0086002 | cardiac muscle cell action potential involved in contraction(GO:0086002) |
| 0.3 | 3.6 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.3 | 0.8 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.2 | 2.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 4.8 | GO:0051098 | regulation of binding(GO:0051098) |
| 0.2 | 2.7 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.2 | 2.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 2.0 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 3.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 1.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 3.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 1.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.2 | 3.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 2.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 2.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.2 | 1.8 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) embryonic skeletal joint development(GO:0072498) |
| 0.2 | 2.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 0.8 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 1.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.0 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.2 | 4.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.2 | 0.6 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 0.9 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 3.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 1.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 1.8 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.1 | 2.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 4.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.4 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 1.9 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 3.2 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 4.4 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 1.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 4.0 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 2.6 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 0.7 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 5.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.5 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 2.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 4.5 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.6 | GO:0071428 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 1.8 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 3.6 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.5 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 1.0 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 3.4 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.5 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.6 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 2.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 3.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.2 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.1 | 1.8 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 2.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 1.1 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.7 | GO:0032324 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 3.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.0 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 2.9 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.1 | 1.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 3.6 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.1 | 2.0 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 2.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.0 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.5 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 2.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 1.2 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 1.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.4 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 8.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 2.5 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 2.8 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 4.4 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 3.7 | GO:0046939 | nucleotide phosphorylation(GO:0046939) |
| 0.0 | 0.8 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 2.6 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 2.1 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 2.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.0 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.3 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.1 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.0 | 1.1 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.2 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 1.6 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.9 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 2.5 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.9 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 1.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.5 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.9 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 2.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 1.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 3.9 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 0.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 1.4 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 2.4 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 2.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.1 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.0 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 2.2 | GO:0030335 | positive regulation of cell migration(GO:0030335) |
| 0.0 | 3.9 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 2.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.8 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 3.4 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 1.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 6.7 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.0 | 1.2 | GO:0000209 | protein polyubiquitination(GO:0000209) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 1.1 | 4.5 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 1.1 | 4.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 1.1 | 5.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.0 | 4.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.8 | 4.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.8 | 2.5 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.8 | 3.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.7 | 2.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.5 | 5.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.5 | 2.5 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.5 | 1.9 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 2.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 2.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.4 | 1.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.4 | 3.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 2.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 2.4 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.3 | 3.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.3 | 7.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 2.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 11.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.3 | 1.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 2.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 0.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 1.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 2.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 2.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 2.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 0.7 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 1.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 3.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 1.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 1.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 1.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 6.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 2.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 4.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.7 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 1.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 2.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 3.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 2.2 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 1.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 3.8 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 8.0 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 1.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 1.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 4.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 3.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.9 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 6.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 154.6 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 1.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 2.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 2.0 | 15.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 1.8 | 5.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.4 | 4.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 1.3 | 8.8 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 1.2 | 5.0 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 1.2 | 7.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.2 | 5.8 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 1.1 | 8.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.9 | 3.8 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.8 | 4.2 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.8 | 4.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.8 | 3.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.8 | 3.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.7 | 5.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.7 | 4.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.7 | 2.2 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.7 | 2.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.7 | 3.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.6 | 2.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.6 | 2.4 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.5 | 2.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.5 | 1.9 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.5 | 1.9 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 2.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.4 | 6.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.4 | 5.8 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.4 | 1.4 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.4 | 1.8 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 2.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 1.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 1.0 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.3 | 2.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 5.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.3 | 4.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 2.7 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.3 | 2.2 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.3 | 4.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 2.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.3 | 0.8 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.2 | 3.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 2.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 12.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 4.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 0.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 2.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 2.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 2.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 1.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 0.7 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.2 | 3.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.2 | 1.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.9 | GO:0097363 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 8.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 4.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.5 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 6.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.4 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 1.1 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 2.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 5.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 8.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 13.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 6.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 2.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.6 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 2.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.9 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.6 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.4 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 1.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 2.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 4.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 3.1 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.2 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 2.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 6.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 15.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.6 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 2.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 3.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 2.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 37.7 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 1.1 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 5.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 4.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.3 | 5.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 13.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 9.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.1 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 3.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 2.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 3.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 9.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.4 | 4.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 8.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.4 | 5.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.4 | 2.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.3 | 6.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.3 | 7.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 5.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 2.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 1.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 2.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 2.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 2.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 9.6 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 0.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 3.8 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 1.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 5.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.0 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 2.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |