Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
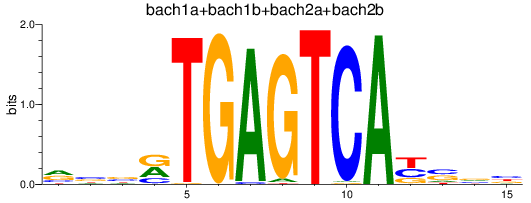
Results for bach1a+bach1b+bach2a+bach2b
Z-value: 0.38
Transcription factors associated with bach1a+bach1b+bach2a+bach2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bach1b
|
ENSDARG00000002196 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
|
bach2b
|
ENSDARG00000004074 | BTB and CNC homology 1, basic leucine zipper transcription factor 2b |
|
bach2a
|
ENSDARG00000036569 | BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
|
bach1a
|
ENSDARG00000062553 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bach1a | dr11_v1_chr15_-_8191992_8191992 | 0.55 | 1.4e-02 | Click! |
| bach1b | dr11_v1_chr10_+_25369254_25369254 | 0.41 | 8.1e-02 | Click! |
| bach2a | dr11_v1_chr17_+_15674052_15674052 | 0.34 | 1.5e-01 | Click! |
| bach2b | dr11_v1_chr20_-_24122881_24122881 | -0.33 | 1.7e-01 | Click! |
Activity profile of bach1a+bach1b+bach2a+bach2b motif
Sorted Z-values of bach1a+bach1b+bach2a+bach2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_618005 | 0.64 |
ENSDART00000189667
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr16_-_24832038 | 0.59 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr4_-_77506362 | 0.59 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr24_+_31361407 | 0.52 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr25_-_11026907 | 0.51 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr10_+_28428222 | 0.50 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr17_+_32623931 | 0.50 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr12_+_25945560 | 0.48 |
ENSDART00000109799
|
mmrn2b
|
multimerin 2b |
| chr11_+_6116096 | 0.46 |
ENSDART00000159680
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr25_-_13408760 | 0.43 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr9_-_48370645 | 0.43 |
ENSDART00000140185
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr2_-_23391266 | 0.37 |
ENSDART00000159048
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr22_-_26595027 | 0.36 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr25_+_18583877 | 0.35 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr18_-_18850302 | 0.33 |
ENSDART00000131965
ENSDART00000167624 |
tgm2l
|
transglutaminase 2, like |
| chr23_-_4705110 | 0.33 |
ENSDART00000144536
ENSDART00000129050 ENSDART00000136399 |
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr4_+_47656992 | 0.33 |
ENSDART00000161148
|
znf1040
|
zinc finger protein 1040 |
| chr8_-_13211527 | 0.32 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr13_-_37122217 | 0.31 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr12_-_27212596 | 0.31 |
ENSDART00000153101
|
psme3
|
proteasome activator subunit 3 |
| chr9_+_22388686 | 0.31 |
ENSDART00000182731
ENSDART00000181462 |
dgkg
|
diacylglycerol kinase, gamma |
| chr12_+_8074343 | 0.31 |
ENSDART00000124084
|
cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr19_+_48359259 | 0.31 |
ENSDART00000167353
|
sgo1
|
shugoshin 1 |
| chr4_+_5317483 | 0.31 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr3_-_15734530 | 0.30 |
ENSDART00000141142
|
mvp
|
major vault protein |
| chr23_-_37113396 | 0.30 |
ENSDART00000102886
ENSDART00000134461 |
zgc:193690
|
zgc:193690 |
| chr20_-_16849306 | 0.30 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr7_-_52963493 | 0.29 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr14_-_493029 | 0.29 |
ENSDART00000115093
|
zgc:172215
|
zgc:172215 |
| chr19_-_27564458 | 0.29 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr10_+_22771176 | 0.28 |
ENSDART00000192046
|
tmem88a
|
transmembrane protein 88 a |
| chr11_+_6116503 | 0.28 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr7_+_14005111 | 0.28 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr23_-_36441693 | 0.28 |
ENSDART00000024354
|
csad
|
cysteine sulfinic acid decarboxylase |
| chr19_+_26072624 | 0.27 |
ENSDART00000147627
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr22_+_35089031 | 0.27 |
ENSDART00000076040
|
srfa
|
serum response factor a |
| chr20_-_34767472 | 0.27 |
ENSDART00000186710
|
paqr8
|
progestin and adipoQ receptor family member VIII |
| chr3_-_15734358 | 0.26 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr12_-_17147473 | 0.26 |
ENSDART00000106035
|
stambpl1
|
STAM binding protein-like 1 |
| chr5_-_66028714 | 0.26 |
ENSDART00000022625
ENSDART00000164228 |
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr11_+_44617021 | 0.26 |
ENSDART00000158887
|
rbm34
|
RNA binding motif protein 34 |
| chr21_+_13233377 | 0.25 |
ENSDART00000142569
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr4_+_16715267 | 0.25 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr12_-_29233738 | 0.25 |
ENSDART00000153175
|
si:ch211-214e3.5
|
si:ch211-214e3.5 |
| chr23_-_4704938 | 0.25 |
ENSDART00000067293
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr24_-_33366188 | 0.24 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr5_+_44805269 | 0.24 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr4_-_56157199 | 0.24 |
ENSDART00000169806
|
si:ch211-207e19.15
|
si:ch211-207e19.15 |
| chr6_-_35309661 | 0.23 |
ENSDART00000114960
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr17_+_51682429 | 0.23 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr14_-_40821411 | 0.23 |
ENSDART00000166621
|
elf1
|
E74-like ETS transcription factor 1 |
| chr7_-_30926030 | 0.23 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr4_-_1801519 | 0.22 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr6_+_56141852 | 0.22 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr4_+_43408004 | 0.22 |
ENSDART00000150476
|
si:dkeyp-53e4.2
|
si:dkeyp-53e4.2 |
| chr25_-_35045250 | 0.21 |
ENSDART00000156508
ENSDART00000126590 |
zgc:114046
|
zgc:114046 |
| chr4_-_60356042 | 0.21 |
ENSDART00000125171
|
znf1121
|
zinc finger protein 1121 |
| chr6_-_18960105 | 0.20 |
ENSDART00000185278
ENSDART00000162968 |
sept9b
|
septin 9b |
| chr15_+_40188076 | 0.20 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr2_+_47945359 | 0.20 |
ENSDART00000098054
|
ftr19
|
finTRIM family, member 19 |
| chr19_-_7495006 | 0.20 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr20_+_2039518 | 0.20 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr2_+_10007113 | 0.20 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr8_+_6576940 | 0.20 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr12_-_3077395 | 0.20 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr13_+_2357637 | 0.20 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr9_-_23181062 | 0.20 |
ENSDART00000192578
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr22_+_24220937 | 0.20 |
ENSDART00000165380
|
uchl5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr13_+_7241170 | 0.19 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr7_+_7696665 | 0.19 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr4_-_4535189 | 0.19 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr13_-_15982707 | 0.19 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr6_-_40842768 | 0.19 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr21_+_22892836 | 0.19 |
ENSDART00000065565
|
alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr12_-_27212880 | 0.19 |
ENSDART00000002835
|
psme3
|
proteasome activator subunit 3 |
| chr3_-_48259289 | 0.19 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr7_+_22688781 | 0.19 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr24_+_38534550 | 0.19 |
ENSDART00000105677
|
zgc:154125
|
zgc:154125 |
| chr10_-_20445549 | 0.19 |
ENSDART00000064613
|
loxl2a
|
lysyl oxidase-like 2a |
| chr20_-_34069956 | 0.19 |
ENSDART00000017941
|
tprb
|
translocated promoter region b, nuclear basket protein |
| chr22_+_36547670 | 0.19 |
ENSDART00000125991
ENSDART00000186372 |
armc9
|
armadillo repeat containing 9 |
| chr5_-_55623443 | 0.19 |
ENSDART00000005671
ENSDART00000176341 |
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr4_+_53183373 | 0.18 |
ENSDART00000176617
|
znf1037
|
zinc finger protein 1037 |
| chr11_+_34760628 | 0.18 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr20_+_42537768 | 0.18 |
ENSDART00000134066
ENSDART00000153434 |
si:dkeyp-93d12.1
|
si:dkeyp-93d12.1 |
| chr1_-_52498146 | 0.18 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr2_+_47944967 | 0.18 |
ENSDART00000135584
|
ftr19
|
finTRIM family, member 19 |
| chr2_+_37837249 | 0.18 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr7_+_42461850 | 0.18 |
ENSDART00000190350
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr8_+_39511932 | 0.18 |
ENSDART00000113511
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr21_-_21020708 | 0.18 |
ENSDART00000064032
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr6_-_21534301 | 0.18 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr17_+_7595356 | 0.17 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr9_-_41025062 | 0.17 |
ENSDART00000002053
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr23_+_24501918 | 0.17 |
ENSDART00000078824
|
szrd1
|
SUZ RNA binding domain containing 1 |
| chr20_-_16548912 | 0.17 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr4_-_17822447 | 0.17 |
ENSDART00000185502
|
dnajb9b
|
DnaJ (Hsp40) homolog, subfamily B, member 9b |
| chr22_-_3595439 | 0.17 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr2_-_3038904 | 0.17 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr14_+_28464736 | 0.17 |
ENSDART00000019003
|
psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr2_-_9544161 | 0.17 |
ENSDART00000124425
|
slc25a24l
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24, like |
| chr21_-_11657043 | 0.17 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr2_-_26642831 | 0.17 |
ENSDART00000132854
ENSDART00000087714 ENSDART00000132651 |
u2surp
|
U2 snRNP-associated SURP domain containing |
| chr11_-_11965033 | 0.17 |
ENSDART00000193683
|
abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr20_-_43723860 | 0.16 |
ENSDART00000122051
|
mixl1
|
Mix paired-like homeobox |
| chr15_-_30450898 | 0.16 |
ENSDART00000156584
|
msi2b
|
musashi RNA-binding protein 2b |
| chr18_+_17537344 | 0.16 |
ENSDART00000025782
|
nup93
|
nucleoporin 93 |
| chr9_-_32668928 | 0.16 |
ENSDART00000135798
|
gzm3.3
|
granzyme 3, tandem duplicate 3 |
| chr2_-_37837472 | 0.16 |
ENSDART00000165347
|
mettl17
|
methyltransferase like 17 |
| chr6_+_33885828 | 0.16 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr16_+_27543893 | 0.16 |
ENSDART00000182421
|
si:ch211-197h24.6
|
si:ch211-197h24.6 |
| chr5_-_63644938 | 0.16 |
ENSDART00000050865
|
surf4l
|
surfeit gene 4, like |
| chr13_-_49585385 | 0.16 |
ENSDART00000165868
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr22_+_39084829 | 0.15 |
ENSDART00000002826
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr18_-_18849916 | 0.15 |
ENSDART00000142422
|
tgm2l
|
transglutaminase 2, like |
| chr25_-_37331513 | 0.15 |
ENSDART00000111862
|
ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr19_-_7272921 | 0.15 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr6_-_43922813 | 0.15 |
ENSDART00000123341
|
prok2
|
prokineticin 2 |
| chr4_+_58057168 | 0.15 |
ENSDART00000161097
|
si:dkey-57k17.1
|
si:dkey-57k17.1 |
| chr19_+_551963 | 0.15 |
ENSDART00000110495
|
akap9
|
A kinase (PRKA) anchor protein 9 |
| chr5_+_44805028 | 0.15 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr5_+_35786141 | 0.14 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr7_-_19638319 | 0.14 |
ENSDART00000163686
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr24_+_38522254 | 0.14 |
ENSDART00000156189
|
si:ch1073-66l23.1
|
si:ch1073-66l23.1 |
| chr11_-_28614608 | 0.14 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr23_+_25856541 | 0.14 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr6_+_56147812 | 0.14 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr7_-_5487593 | 0.14 |
ENSDART00000136594
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr4_+_68087932 | 0.14 |
ENSDART00000150494
|
znf1096
|
zinc finger protein 1096 |
| chr5_-_42950963 | 0.14 |
ENSDART00000149868
|
grsf1
|
G-rich RNA sequence binding factor 1 |
| chr16_+_42829735 | 0.14 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr5_-_14564878 | 0.14 |
ENSDART00000160511
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr15_-_23442891 | 0.14 |
ENSDART00000059376
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr20_+_6543674 | 0.14 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr11_+_807153 | 0.14 |
ENSDART00000173289
|
vgll4b
|
vestigial-like family member 4b |
| chr21_+_26620867 | 0.13 |
ENSDART00000176008
|
esrra
|
estrogen-related receptor alpha |
| chr11_+_45219558 | 0.13 |
ENSDART00000167828
|
tmc6b
|
transmembrane channel-like 6b |
| chr1_-_52497834 | 0.13 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr14_-_17588345 | 0.13 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr18_-_37355666 | 0.12 |
ENSDART00000098914
|
yap1
|
Yes-associated protein 1 |
| chr5_-_33298352 | 0.12 |
ENSDART00000015076
|
zgc:103692
|
zgc:103692 |
| chr23_+_17522867 | 0.12 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr24_-_21973365 | 0.12 |
ENSDART00000081204
ENSDART00000030592 |
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| chr5_+_20035284 | 0.12 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr23_+_7548797 | 0.12 |
ENSDART00000006765
|
tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr1_+_52137528 | 0.12 |
ENSDART00000007079
ENSDART00000074265 |
asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr4_+_9508505 | 0.12 |
ENSDART00000080842
|
kitlgb
|
kit ligand b |
| chr3_-_16493528 | 0.12 |
ENSDART00000142099
|
si:ch211-23l10.3
|
si:ch211-23l10.3 |
| chr3_-_3448095 | 0.12 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr13_-_51903150 | 0.12 |
ENSDART00000090644
|
mrpl2
|
mitochondrial ribosomal protein L2 |
| chr22_-_11724563 | 0.12 |
ENSDART00000190796
ENSDART00000184744 |
krt222
|
keratin 222 |
| chr25_-_7753207 | 0.12 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr24_+_80653 | 0.11 |
ENSDART00000158473
ENSDART00000129135 |
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr24_-_5691956 | 0.11 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr4_+_25181572 | 0.11 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr9_+_28140089 | 0.11 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr15_-_36727462 | 0.11 |
ENSDART00000085971
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr22_+_39084107 | 0.11 |
ENSDART00000191346
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr18_+_7543347 | 0.11 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr15_+_17343319 | 0.11 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr5_+_9037650 | 0.11 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr3_+_29476085 | 0.10 |
ENSDART00000184495
ENSDART00000181058 |
fam83fa
|
family with sequence similarity 83, member Fa |
| chr8_-_38201415 | 0.10 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr22_-_6562618 | 0.10 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr23_+_45200481 | 0.10 |
ENSDART00000004357
ENSDART00000111126 ENSDART00000193560 ENSDART00000190476 |
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr4_+_64147241 | 0.10 |
ENSDART00000163509
|
znf1089
|
zinc finger protein 1089 |
| chr2_-_37837056 | 0.10 |
ENSDART00000158179
ENSDART00000160317 ENSDART00000171409 |
mettl17
|
methyltransferase like 17 |
| chr5_+_37785152 | 0.10 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr7_+_26649319 | 0.10 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr8_+_48966165 | 0.10 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr4_+_69624049 | 0.10 |
ENSDART00000170052
|
znf1030
|
zinc finger protein 1030 |
| chr6_-_43233700 | 0.10 |
ENSDART00000113706
|
trnt1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr5_+_37087583 | 0.10 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr8_+_48965767 | 0.10 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr4_+_40063572 | 0.10 |
ENSDART00000146710
|
BX649521.1
|
|
| chr4_+_76431531 | 0.09 |
ENSDART00000174278
|
FP074874.1
|
|
| chr20_-_32446406 | 0.09 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr21_+_25765734 | 0.09 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr5_+_34549845 | 0.09 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr11_-_39118882 | 0.09 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr5_+_44804791 | 0.09 |
ENSDART00000122288
|
ctsla
|
cathepsin La |
| chr19_+_37120491 | 0.09 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr24_+_42074143 | 0.09 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr1_-_10577945 | 0.09 |
ENSDART00000179237
ENSDART00000040502 ENSDART00000186876 |
trpc5a
|
transient receptor potential cation channel, subfamily C, member 5a |
| chr12_+_20336070 | 0.09 |
ENSDART00000066385
|
zgc:163057
|
zgc:163057 |
| chr2_+_25278107 | 0.09 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr13_+_43247936 | 0.09 |
ENSDART00000126850
ENSDART00000165331 |
smoc2
|
SPARC related modular calcium binding 2 |
| chr21_+_10866421 | 0.09 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr7_-_5396154 | 0.09 |
ENSDART00000172980
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr24_-_31090948 | 0.09 |
ENSDART00000176799
|
hccsb
|
holocytochrome c synthase b |
| chr4_-_71708567 | 0.09 |
ENSDART00000182645
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr18_-_8396192 | 0.09 |
ENSDART00000193830
|
BEND7
|
si:ch211-220f12.4 |
| chr9_+_41024973 | 0.09 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr23_-_45339439 | 0.08 |
ENSDART00000148726
|
ccdc171
|
coiled-coil domain containing 171 |
| chr17_+_28675120 | 0.08 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
| chr7_+_34549198 | 0.08 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr5_-_49951106 | 0.08 |
ENSDART00000135954
|
fam172a
|
family with sequence similarity 172, member A |
| chr23_-_28239750 | 0.08 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr8_-_34402839 | 0.08 |
ENSDART00000180173
ENSDART00000191588 ENSDART00000186376 |
gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr11_-_22371105 | 0.08 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
Network of associatons between targets according to the STRING database.
First level regulatory network of bach1a+bach1b+bach2a+bach2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.3 | GO:1901295 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.5 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.3 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.1 | 0.3 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.4 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.4 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.4 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.0 | 0.2 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.2 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.3 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.0 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.1 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.5 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.6 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.2 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.3 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |