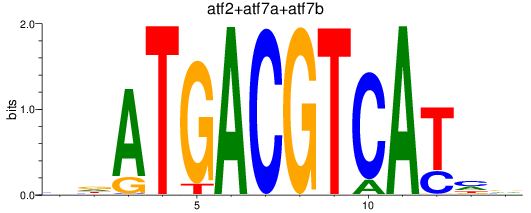
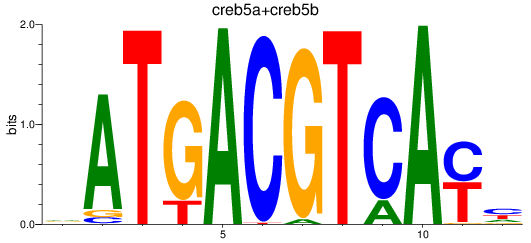
Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for atf2+atf7a+atf7b_creb5a+creb5b
Z-value: 0.83
Transcription factors associated with atf2+atf7a+atf7b_creb5a+creb5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf7a
|
ENSDARG00000011298 | activating transcription factor 7a |
|
atf2
|
ENSDARG00000023903 | activating transcription factor 2 |
|
atf7b
|
ENSDARG00000055481 | activating transcription factor 7b |
|
atf7b
|
ENSDARG00000114492 | activating transcription factor 7b |
|
atf7b
|
ENSDARG00000115171 | activating transcription factor 7b |
|
creb5b
|
ENSDARG00000070536 | cAMP responsive element binding protein 5b |
|
creb5a
|
ENSDARG00000099002 | cAMP responsive element binding protein 5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf7a | dr11_v1_chr23_-_27479558_27479558 | 0.50 | 2.9e-02 | Click! |
| atf7b | dr11_v1_chr6_-_39631164_39631181 | 0.38 | 1.1e-01 | Click! |
| atf2 | dr11_v1_chr9_+_2343096_2343172 | 0.33 | 1.7e-01 | Click! |
| creb5a | dr11_v1_chr19_-_19505167_19505167 | 0.15 | 5.3e-01 | Click! |
| creb5b | dr11_v1_chr16_-_20707742_20707742 | -0.04 | 8.7e-01 | Click! |
Activity profile of atf2+atf7a+atf7b_creb5a+creb5b motif
Sorted Z-values of atf2+atf7a+atf7b_creb5a+creb5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_33121535 | 1.13 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr22_+_1911269 | 1.06 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr15_+_28685625 | 0.90 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr15_+_28685892 | 0.88 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr7_-_12065668 | 0.87 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr5_+_69808763 | 0.81 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr5_+_56268436 | 0.79 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr24_+_4373355 | 0.69 |
ENSDART00000179062
ENSDART00000093256 ENSDART00000138943 |
ccny
|
cyclin Y |
| chr4_+_77060861 | 0.68 |
ENSDART00000174271
ENSDART00000174393 ENSDART00000150450 |
si:dkey-240n22.8
|
si:dkey-240n22.8 |
| chr20_-_24122881 | 0.67 |
ENSDART00000131857
|
bach2b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2b |
| chr9_-_306515 | 0.67 |
ENSDART00000166059
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr19_-_1961024 | 0.65 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr1_-_44037843 | 0.63 |
ENSDART00000160276
|
si:ch73-109d9.3
|
si:ch73-109d9.3 |
| chr22_+_2315996 | 0.61 |
ENSDART00000132489
|
znf1175
|
zinc finger protein 1175 |
| chr7_+_7552008 | 0.60 |
ENSDART00000173018
ENSDART00000049311 |
clcn3
|
chloride channel 3 |
| chr3_+_53116172 | 0.57 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr16_-_32649929 | 0.57 |
ENSDART00000136161
|
faxcb
|
failed axon connections homolog b |
| chr20_-_32446406 | 0.57 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr23_+_34990693 | 0.56 |
ENSDART00000013449
|
CHST13
|
si:ch211-236h17.3 |
| chr3_-_5964557 | 0.55 |
ENSDART00000184738
|
BX284638.1
|
|
| chr4_+_77141970 | 0.54 |
ENSDART00000174209
|
si:dkey-172k15.11
|
si:dkey-172k15.11 |
| chr2_+_58877162 | 0.53 |
ENSDART00000122174
|
CABZ01085658.1
|
|
| chr22_+_38935060 | 0.53 |
ENSDART00000183732
ENSDART00000130055 |
sirt7
|
sirtuin 7 |
| chr3_-_16413606 | 0.52 |
ENSDART00000127309
ENSDART00000017172 ENSDART00000136465 |
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr23_-_25686894 | 0.52 |
ENSDART00000181420
ENSDART00000088208 |
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr4_+_77151419 | 0.52 |
ENSDART00000174312
|
CU467646.4
|
|
| chr24_+_39638555 | 0.51 |
ENSDART00000078313
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr3_-_49566364 | 0.51 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr4_-_64482414 | 0.51 |
ENSDART00000159000
|
znf1105
|
zinc finger protein 1105 |
| chr17_+_23554932 | 0.50 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr15_-_34658057 | 0.50 |
ENSDART00000110964
|
bag6
|
BCL2 associated athanogene 6 |
| chr22_+_30330574 | 0.50 |
ENSDART00000104751
|
mxi1
|
max interactor 1, dimerization protein |
| chr20_-_37933237 | 0.49 |
ENSDART00000142567
ENSDART00000036371 ENSDART00000061445 |
angel2
|
angel homolog 2 (Drosophila) |
| chr22_+_2183110 | 0.46 |
ENSDART00000159279
ENSDART00000121703 |
znf1152
|
zinc finger protein 1152 |
| chr24_-_19718077 | 0.45 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr9_+_219124 | 0.45 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr20_+_29743904 | 0.45 |
ENSDART00000146366
ENSDART00000153154 |
kidins220b
|
kinase D-interacting substrate 220b |
| chr1_-_23595779 | 0.44 |
ENSDART00000134860
ENSDART00000138852 |
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr8_+_29742237 | 0.43 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr21_-_23308286 | 0.42 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr16_+_34523515 | 0.41 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr16_+_25137483 | 0.41 |
ENSDART00000155666
|
znf576.1
|
zinc finger protein 576, tandem duplicate 1 |
| chr25_+_19201231 | 0.41 |
ENSDART00000067323
|
hapln3
|
hyaluronan and proteoglycan link protein 3 |
| chr4_-_43640507 | 0.41 |
ENSDART00000150700
|
si:dkey-29p23.2
|
si:dkey-29p23.2 |
| chr13_-_865193 | 0.41 |
ENSDART00000187053
|
AL929536.7
|
|
| chr15_+_618376 | 0.40 |
ENSDART00000156007
|
si:ch73-144d13.8
|
si:ch73-144d13.8 |
| chr19_+_2279051 | 0.39 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr21_+_45816030 | 0.39 |
ENSDART00000187056
|
pitx1
|
paired-like homeodomain 1 |
| chr16_-_33097398 | 0.39 |
ENSDART00000166617
|
dopey1
|
dopey family member 1 |
| chr8_-_410199 | 0.37 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr20_-_9760424 | 0.37 |
ENSDART00000104936
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr4_+_39055027 | 0.37 |
ENSDART00000150397
|
znf977
|
zinc finger protein 977 |
| chr6_-_29049022 | 0.37 |
ENSDART00000190309
|
evi5b
|
ecotropic viral integration site 5b |
| chr22_+_2239254 | 0.37 |
ENSDART00000131396
ENSDART00000135320 |
znf1144
|
zinc finger protein 1144 |
| chr20_-_3319642 | 0.37 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr15_+_872085 | 0.37 |
ENSDART00000193867
ENSDART00000157483 ENSDART00000156324 |
si:dkey-7i4.9
si:dkey-77f5.13
si:dkey-7i4.11
|
si:dkey-7i4.9 si:dkey-77f5.13 si:dkey-7i4.11 |
| chr18_-_20444296 | 0.37 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr4_+_30775376 | 0.36 |
ENSDART00000158528
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr19_-_7225060 | 0.36 |
ENSDART00000133179
ENSDART00000151138 ENSDART00000104799 ENSDART00000128331 |
col11a2
|
collagen, type XI, alpha 2 |
| chr22_+_2254972 | 0.36 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr8_-_24113575 | 0.36 |
ENSDART00000099692
ENSDART00000186211 |
dclre1b
|
DNA cross-link repair 1B |
| chr21_-_5007109 | 0.35 |
ENSDART00000187042
ENSDART00000097796 ENSDART00000146766 |
rnf165a
|
ring finger protein 165a |
| chr15_+_879897 | 0.35 |
ENSDART00000185066
ENSDART00000179840 |
si:dkey-7i4.9
si:dkey-77f5.13
|
si:dkey-7i4.9 si:dkey-77f5.13 |
| chr4_-_36144500 | 0.35 |
ENSDART00000170896
|
znf992
|
zinc finger protein 992 |
| chr9_-_296169 | 0.35 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr25_-_6447835 | 0.34 |
ENSDART00000012820
|
snupn
|
snurportin 1 |
| chr16_+_5184402 | 0.33 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr21_+_19547806 | 0.33 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr4_+_40953320 | 0.33 |
ENSDART00000151912
|
znf1136
|
zinc finger protein 1136 |
| chr5_-_30145939 | 0.33 |
ENSDART00000086795
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr7_+_34549198 | 0.33 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr3_-_2593859 | 0.33 |
ENSDART00000143826
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr15_-_27710513 | 0.33 |
ENSDART00000005641
ENSDART00000134373 |
lhx1a
|
LIM homeobox 1a |
| chr22_+_1821718 | 0.32 |
ENSDART00000132220
|
znf1002
|
zinc finger protein 1002 |
| chr3_+_20156956 | 0.32 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr22_+_997838 | 0.32 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr7_+_40228422 | 0.32 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr4_+_359970 | 0.32 |
ENSDART00000139832
|
tmem181
|
transmembrane protein 181 |
| chr16_-_41131578 | 0.32 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr9_-_28939181 | 0.31 |
ENSDART00000101276
ENSDART00000135334 |
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr5_-_23999777 | 0.31 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr7_+_24889783 | 0.31 |
ENSDART00000005329
ENSDART00000159955 |
mark2b
|
MAP/microtubule affinity-regulating kinase 2b |
| chr13_+_28705143 | 0.31 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr5_+_9047433 | 0.31 |
ENSDART00000091463
|
idua
|
iduronidase, alpha-L- |
| chr4_-_73739119 | 0.30 |
ENSDART00000108669
|
zgc:171551
|
zgc:171551 |
| chr11_-_44163164 | 0.30 |
ENSDART00000047126
|
clcn4
|
chloride channel, voltage-sensitive 4 |
| chr20_-_54245256 | 0.30 |
ENSDART00000170482
|
prpf39
|
PRP39 pre-mRNA processing factor 39 homolog (yeast) |
| chr4_-_6373735 | 0.30 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr9_+_7548533 | 0.30 |
ENSDART00000081543
|
ptprna
|
protein tyrosine phosphatase, receptor type, Na |
| chr2_-_43739559 | 0.29 |
ENSDART00000138947
|
kif5ba
|
kinesin family member 5B, a |
| chr10_-_11353101 | 0.29 |
ENSDART00000092110
|
lin54
|
lin-54 DREAM MuvB core complex component |
| chr18_-_46063773 | 0.29 |
ENSDART00000078561
|
si:ch73-262h23.4
|
si:ch73-262h23.4 |
| chr4_+_76304911 | 0.29 |
ENSDART00000172734
ENSDART00000161850 |
si:ch73-389k6.1
|
si:ch73-389k6.1 |
| chr1_-_18592068 | 0.29 |
ENSDART00000082063
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr24_-_41657005 | 0.29 |
ENSDART00000159109
|
CABZ01044099.1
|
|
| chr25_+_16945348 | 0.28 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr13_-_3155243 | 0.28 |
ENSDART00000139183
ENSDART00000050934 |
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr5_-_19444930 | 0.28 |
ENSDART00000136259
ENSDART00000188499 |
kctd10
|
potassium channel tetramerization domain containing 10 |
| chr1_-_23596391 | 0.28 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr18_+_12058403 | 0.28 |
ENSDART00000140854
ENSDART00000193632 ENSDART00000190519 ENSDART00000190685 ENSDART00000112671 |
bicd1a
|
bicaudal D homolog 1a |
| chr1_-_11291324 | 0.28 |
ENSDART00000091205
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr11_+_28166165 | 0.28 |
ENSDART00000169360
ENSDART00000192311 |
ephb2b
|
eph receptor B2b |
| chr12_-_10365875 | 0.28 |
ENSDART00000142386
ENSDART00000007335 |
ndc80
|
NDC80 kinetochore complex component |
| chr8_+_1839695 | 0.28 |
ENSDART00000148254
ENSDART00000143473 |
snap29
|
synaptosomal-associated protein 29 |
| chr5_-_32363372 | 0.28 |
ENSDART00000098045
|
gas1b
|
growth arrest-specific 1b |
| chr15_-_5892650 | 0.28 |
ENSDART00000154812
|
brwd1
|
bromodomain and WD repeat domain containing 1 |
| chr25_-_6448050 | 0.27 |
ENSDART00000180616
|
snupn
|
snurportin 1 |
| chr4_-_42294516 | 0.27 |
ENSDART00000133558
|
si:dkey-4e4.1
|
si:dkey-4e4.1 |
| chr10_-_2971407 | 0.27 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr17_-_24866727 | 0.27 |
ENSDART00000027957
|
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr21_-_44772738 | 0.27 |
ENSDART00000026178
|
kif4
|
kinesin family member 4 |
| chr20_+_46699021 | 0.27 |
ENSDART00000167398
ENSDART00000029894 |
wdr26b
|
WD repeat domain 26b |
| chr4_-_73825089 | 0.27 |
ENSDART00000174207
|
si:dkey-262g12.12
|
si:dkey-262g12.12 |
| chr22_+_2819613 | 0.27 |
ENSDART00000131234
|
si:dkey-20i20.3
|
si:dkey-20i20.3 |
| chr1_-_17693273 | 0.26 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr4_-_43056627 | 0.26 |
ENSDART00000150269
|
si:dkey-54j5.2
|
si:dkey-54j5.2 |
| chr9_-_6502491 | 0.26 |
ENSDART00000102672
|
nck2a
|
NCK adaptor protein 2a |
| chr13_+_47623916 | 0.26 |
ENSDART00000109266
|
mertka
|
c-mer proto-oncogene tyrosine kinase a |
| chr1_+_19538299 | 0.26 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr4_+_48769555 | 0.26 |
ENSDART00000150725
|
znf1020
|
zinc finger protein 1020 |
| chr5_+_22133153 | 0.25 |
ENSDART00000016214
|
msna
|
moesin a |
| chr1_-_9940494 | 0.25 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr4_+_76403698 | 0.25 |
ENSDART00000184821
ENSDART00000169373 |
FP074874.2
FP074874.1
|
|
| chr15_+_1796313 | 0.25 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr4_+_45504471 | 0.24 |
ENSDART00000150399
|
si:dkey-256i11.2
|
si:dkey-256i11.2 |
| chr15_+_17030473 | 0.24 |
ENSDART00000129407
|
plin2
|
perilipin 2 |
| chr12_+_36109507 | 0.24 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr23_+_43424518 | 0.24 |
ENSDART00000022498
|
tti1
|
TELO2 interacting protein 1 |
| chr4_+_65332578 | 0.24 |
ENSDART00000170824
|
znf1108
|
zinc finger protein 1108 |
| chr22_+_2228919 | 0.24 |
ENSDART00000133475
|
znf1161
|
zinc finger protein 1161 |
| chr17_-_32621103 | 0.24 |
ENSDART00000155321
|
xkr5b
|
XK related 5b |
| chr5_-_32882162 | 0.24 |
ENSDART00000085769
|
lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr10_-_11353469 | 0.24 |
ENSDART00000191847
|
lin54
|
lin-54 DREAM MuvB core complex component |
| chr6_+_58832323 | 0.24 |
ENSDART00000042595
|
dctn2
|
dynactin 2 (p50) |
| chr12_-_5455936 | 0.24 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr12_+_33894396 | 0.24 |
ENSDART00000130853
ENSDART00000152988 |
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr4_-_149334 | 0.24 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr5_+_28259655 | 0.23 |
ENSDART00000087684
|
ncaph
|
non-SMC condensin I complex, subunit H |
| chr25_+_34845469 | 0.23 |
ENSDART00000145416
|
tmem231
|
transmembrane protein 231 |
| chr1_-_38756870 | 0.23 |
ENSDART00000130324
ENSDART00000148404 |
gpm6ab
|
glycoprotein M6Ab |
| chr13_+_22295905 | 0.23 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr15_-_762319 | 0.23 |
ENSDART00000154306
ENSDART00000157492 |
si:dkey-7i4.16
znf1011
|
si:dkey-7i4.16 zinc finger protein 1011 |
| chr2_-_30324610 | 0.23 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr22_+_1947494 | 0.23 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr13_+_51710725 | 0.23 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr20_+_25911342 | 0.23 |
ENSDART00000146004
|
ttbk2b
|
tau tubulin kinase 2b |
| chr22_-_18746508 | 0.23 |
ENSDART00000003929
|
midn
|
midnolin |
| chr21_-_45086170 | 0.23 |
ENSDART00000188963
|
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr5_-_31856681 | 0.23 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr4_-_44801146 | 0.22 |
ENSDART00000154874
|
si:dkeyp-77c4.5
|
si:dkeyp-77c4.5 |
| chr2_-_17115256 | 0.22 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr2_-_17114852 | 0.22 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr2_-_42415902 | 0.22 |
ENSDART00000142489
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr19_-_26923273 | 0.22 |
ENSDART00000089609
|
skiv2l
|
SKI2 homolog, superkiller viralicidic activity 2-like |
| chr4_-_48957129 | 0.22 |
ENSDART00000150426
|
znf1064
|
zinc finger protein 1064 |
| chr10_-_41906609 | 0.22 |
ENSDART00000102530
|
kdm2bb
|
lysine (K)-specific demethylase 2Bb |
| chr21_-_43606502 | 0.22 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr5_+_51848756 | 0.22 |
ENSDART00000087467
ENSDART00000184466 |
cmya5
|
cardiomyopathy associated 5 |
| chr15_-_23443937 | 0.22 |
ENSDART00000148311
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr23_-_4409668 | 0.22 |
ENSDART00000081823
|
si:ch73-142c19.1
|
si:ch73-142c19.1 |
| chr15_+_903743 | 0.21 |
ENSDART00000155783
|
si:dkey-77f5.13
|
si:dkey-77f5.13 |
| chr21_+_45366229 | 0.21 |
ENSDART00000029946
|
ube2b
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog) |
| chr25_-_32751982 | 0.21 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr11_-_575786 | 0.21 |
ENSDART00000019997
|
mkrn2
|
makorin, ring finger protein, 2 |
| chr25_+_3104959 | 0.21 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr25_+_35683956 | 0.21 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr4_+_77076645 | 0.21 |
ENSDART00000174346
|
BX005417.2
|
|
| chr1_+_52792439 | 0.21 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr20_-_28352352 | 0.21 |
ENSDART00000128806
|
ino80
|
INO80 complex subunit |
| chr10_-_17170086 | 0.21 |
ENSDART00000020122
|
ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr10_-_1276046 | 0.21 |
ENSDART00000169779
|
pdlim5b
|
PDZ and LIM domain 5b |
| chr23_-_18981595 | 0.21 |
ENSDART00000147617
|
bcl2l1
|
bcl2-like 1 |
| chr15_+_618081 | 0.20 |
ENSDART00000181518
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr4_-_41825115 | 0.20 |
ENSDART00000160196
|
si:dkey-237m9.1
|
si:dkey-237m9.1 |
| chr4_-_57869437 | 0.20 |
ENSDART00000170067
|
zgc:174314
|
zgc:174314 |
| chr16_-_32208728 | 0.20 |
ENSDART00000023995
|
calhm5.2
|
calcium homeostasis modulator family member 5, tandem duplicate 2 |
| chr2_-_10386738 | 0.20 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr5_-_41494831 | 0.20 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr21_-_435466 | 0.20 |
ENSDART00000110297
|
klf4
|
Kruppel-like factor 4 |
| chr4_-_73448588 | 0.20 |
ENSDART00000138333
|
si:ch73-120g24.4
|
si:ch73-120g24.4 |
| chr23_+_10805188 | 0.20 |
ENSDART00000035693
|
ppp4r2a
|
protein phosphatase 4, regulatory subunit 2a |
| chr9_-_370225 | 0.19 |
ENSDART00000163637
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr10_-_26273629 | 0.19 |
ENSDART00000147790
|
dchs1b
|
dachsous cadherin-related 1b |
| chr14_-_38946808 | 0.19 |
ENSDART00000139293
|
gla
|
galactosidase, alpha |
| chr4_-_33405483 | 0.19 |
ENSDART00000150625
|
CT027578.1
|
|
| chr3_-_1204341 | 0.19 |
ENSDART00000089646
|
fam234b
|
family with sequence similarity 234, member B |
| chr8_+_21406769 | 0.19 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr12_+_48204891 | 0.19 |
ENSDART00000190534
ENSDART00000164427 |
ndr2
|
nodal-related 2 |
| chr1_+_9290103 | 0.19 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr2_-_43739740 | 0.19 |
ENSDART00000113849
|
kif5ba
|
kinesin family member 5B, a |
| chr15_-_780472 | 0.19 |
ENSDART00000188131
ENSDART00000156942 |
si:dkey-7i4.21
si:dkey-7i4.15
|
si:dkey-7i4.21 si:dkey-7i4.15 |
| chr17_-_19345521 | 0.19 |
ENSDART00000082085
|
gsc
|
goosecoid |
| chr23_-_35396845 | 0.19 |
ENSDART00000142038
ENSDART00000049373 ENSDART00000181978 ENSDART00000171357 |
cmtr1
|
cap methyltransferase 1 |
| chr6_-_48311 | 0.19 |
ENSDART00000131010
|
zgc:114175
|
zgc:114175 |
| chr5_-_24000211 | 0.19 |
ENSDART00000188865
|
map7d2a
|
MAP7 domain containing 2a |
| chr4_-_30269748 | 0.18 |
ENSDART00000163550
|
si:dkey-233e3.3
|
si:dkey-233e3.3 |
| chr1_-_55068941 | 0.18 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr8_-_38317914 | 0.18 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr10_-_26274094 | 0.18 |
ENSDART00000108798
|
dchs1b
|
dachsous cadherin-related 1b |
| chr22_+_2660505 | 0.18 |
ENSDART00000135555
ENSDART00000031485 |
znf1162
|
zinc finger protein 1162 |
Network of associatons between targets according to the STRING database.
First level regulatory network of atf2+atf7a+atf7b_creb5a+creb5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 0.6 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.1 | 0.7 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.6 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.4 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.1 | 0.8 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.3 | GO:0097377 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.1 | 0.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.3 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.4 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.1 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.2 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.5 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.3 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.1 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.2 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.2 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.3 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.0 | 0.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.4 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.0 | 0.1 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 1.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.2 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.2 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.2 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.5 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.1 | 0.4 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 0.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.1 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.0 | 0.1 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 1.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.0 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |