Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
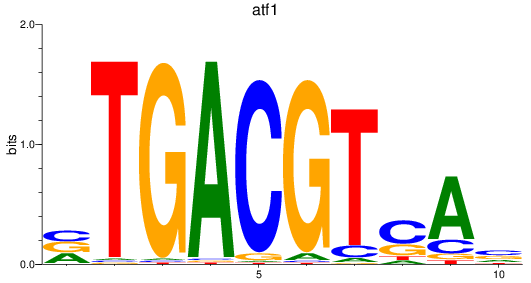
Results for atf1
Z-value: 1.65
Transcription factors associated with atf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf1
|
ENSDARG00000044301 | activating transcription factor 1 |
|
atf1
|
ENSDARG00000109865 | activating transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf1 | dr11_v1_chr6_-_39518489_39518489 | 0.91 | 9.0e-08 | Click! |
Activity profile of atf1 motif
Sorted Z-values of atf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_11685742 | 5.64 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr20_+_24448007 | 4.48 |
ENSDART00000139866
|
si:dkey-273g18.1
|
si:dkey-273g18.1 |
| chr21_-_23308286 | 4.11 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr16_-_42013858 | 3.60 |
ENSDART00000045403
|
etv2
|
ets variant 2 |
| chr11_-_41996957 | 3.49 |
ENSDART00000055706
|
her15.2
|
hairy and enhancer of split-related 15, tandem duplicate 2 |
| chr5_+_36693859 | 3.42 |
ENSDART00000019259
|
dlb
|
deltaB |
| chr21_-_23307653 | 3.38 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr25_+_34845115 | 3.08 |
ENSDART00000061996
|
tmem231
|
transmembrane protein 231 |
| chr2_+_12255568 | 2.97 |
ENSDART00000184164
ENSDART00000013454 |
prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr18_+_619619 | 2.70 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr22_+_1911269 | 2.68 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr25_+_34845469 | 2.63 |
ENSDART00000145416
|
tmem231
|
transmembrane protein 231 |
| chr16_-_42014272 | 2.60 |
ENSDART00000180488
|
etv2
|
ets variant 2 |
| chr19_-_18418763 | 2.58 |
ENSDART00000167271
|
zgc:112966
|
zgc:112966 |
| chr7_-_12065668 | 2.57 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr7_-_54677143 | 2.55 |
ENSDART00000163748
|
ccnd1
|
cyclin D1 |
| chr23_+_44634187 | 2.45 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr16_+_52966812 | 2.42 |
ENSDART00000148435
ENSDART00000049099 ENSDART00000150117 |
trip13
|
thyroid hormone receptor interactor 13 |
| chr1_-_20068155 | 2.36 |
ENSDART00000102993
|
mettl14
|
methyltransferase like 14 |
| chr2_-_36674005 | 2.32 |
ENSDART00000004899
|
ccnl1b
|
cyclin L1b |
| chr22_-_10541372 | 2.32 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr1_+_10110203 | 2.32 |
ENSDART00000080576
ENSDART00000181437 |
lratb.1
|
lecithin retinol acyltransferase b, tandem duplicate 1 |
| chr4_-_57001458 | 2.31 |
ENSDART00000158660
|
si:ch211-161m3.1
|
si:ch211-161m3.1 |
| chr16_-_41535690 | 2.29 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr7_+_1579236 | 2.28 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr5_-_32363372 | 2.27 |
ENSDART00000098045
|
gas1b
|
growth arrest-specific 1b |
| chr12_-_10365875 | 2.26 |
ENSDART00000142386
ENSDART00000007335 |
ndc80
|
NDC80 kinetochore complex component |
| chr4_-_25515154 | 2.26 |
ENSDART00000186524
|
rbm17
|
RNA binding motif protein 17 |
| chr4_+_26036682 | 2.26 |
ENSDART00000126474
|
si:ch211-265o23.1
|
si:ch211-265o23.1 |
| chr5_+_43870389 | 2.25 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr22_+_336256 | 2.23 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr15_+_36309070 | 2.23 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr5_+_9047433 | 2.22 |
ENSDART00000091463
|
idua
|
iduronidase, alpha-L- |
| chr1_-_18592068 | 2.20 |
ENSDART00000082063
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr17_+_32623931 | 2.18 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr20_-_32446406 | 2.18 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr22_+_20135443 | 2.17 |
ENSDART00000143641
|
eef2a.1
|
eukaryotic translation elongation factor 2a, tandem duplicate 1 |
| chr11_+_42494531 | 2.17 |
ENSDART00000067604
|
arf4a
|
ADP-ribosylation factor 4a |
| chr1_-_17693273 | 2.17 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr14_+_39156 | 2.15 |
ENSDART00000082184
|
tmem107
|
transmembrane protein 107 |
| chr10_+_16165533 | 2.14 |
ENSDART00000065045
|
prrc1
|
proline-rich coiled-coil 1 |
| chr1_+_52792439 | 2.12 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr23_+_36144487 | 2.12 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr4_+_18441988 | 2.12 |
ENSDART00000040827
|
ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr17_+_37932706 | 2.11 |
ENSDART00000075941
|
plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr23_+_32011768 | 2.10 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr4_+_45504471 | 2.07 |
ENSDART00000150399
|
si:dkey-256i11.2
|
si:dkey-256i11.2 |
| chr5_-_32505109 | 2.07 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr2_-_10386738 | 2.06 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr9_-_27749936 | 2.06 |
ENSDART00000064156
|
tbccd1
|
TBCC domain containing 1 |
| chr7_-_48667056 | 2.05 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr14_-_31619408 | 2.03 |
ENSDART00000173277
|
mmgt1
|
membrane magnesium transporter 1 |
| chr13_+_47623916 | 2.03 |
ENSDART00000109266
|
mertka
|
c-mer proto-oncogene tyrosine kinase a |
| chr9_+_34148714 | 2.03 |
ENSDART00000078051
|
gpr161
|
G protein-coupled receptor 161 |
| chr5_-_32505276 | 2.02 |
ENSDART00000034705
ENSDART00000187597 |
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr23_+_28718863 | 2.00 |
ENSDART00000078156
|
srm
|
spermidine synthase |
| chr13_+_46941930 | 1.99 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr15_-_34658057 | 1.98 |
ENSDART00000110964
|
bag6
|
BCL2 associated athanogene 6 |
| chr13_-_23612324 | 1.97 |
ENSDART00000136406
ENSDART00000005004 |
prim2
|
DNA primase subunit 2 |
| chr15_-_18361475 | 1.97 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr17_+_15535501 | 1.96 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr13_-_13030851 | 1.95 |
ENSDART00000009499
|
nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr4_+_42556555 | 1.95 |
ENSDART00000168536
|
znf1053
|
zinc finger protein 1053 |
| chr18_-_30499489 | 1.94 |
ENSDART00000033746
|
gins2
|
GINS complex subunit 2 |
| chr18_+_40381102 | 1.94 |
ENSDART00000136588
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr25_-_6447835 | 1.91 |
ENSDART00000012820
|
snupn
|
snurportin 1 |
| chr1_+_19538299 | 1.90 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr25_-_32363341 | 1.89 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr7_+_34549198 | 1.89 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr22_+_1947494 | 1.88 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr5_+_24086227 | 1.86 |
ENSDART00000051549
ENSDART00000177458 ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr15_+_23443665 | 1.85 |
ENSDART00000059369
|
phykpl
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr7_+_69449814 | 1.85 |
ENSDART00000109644
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr22_+_12162470 | 1.85 |
ENSDART00000053045
ENSDART00000143599 |
ccnt2b
|
cyclin T2b |
| chr24_+_39638555 | 1.84 |
ENSDART00000078313
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr7_+_66634167 | 1.84 |
ENSDART00000027616
|
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr7_+_39166460 | 1.81 |
ENSDART00000052318
ENSDART00000146635 ENSDART00000173877 ENSDART00000173767 ENSDART00000173600 |
mdka
|
midkine a |
| chr22_-_15593824 | 1.81 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr11_+_36379293 | 1.80 |
ENSDART00000135360
|
atxn7l2a
|
ataxin 7-like 2a |
| chr11_+_31380495 | 1.79 |
ENSDART00000185073
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr6_-_32093830 | 1.79 |
ENSDART00000017695
|
foxd3
|
forkhead box D3 |
| chr16_-_21915223 | 1.78 |
ENSDART00000170634
|
setdb1b
|
SET domain, bifurcated 1b |
| chr24_+_31361407 | 1.78 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr22_-_10541712 | 1.78 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr3_-_62194512 | 1.76 |
ENSDART00000074174
|
tbl3
|
transducin (beta)-like 3 |
| chr11_-_27827442 | 1.74 |
ENSDART00000121847
ENSDART00000132018 ENSDART00000145744 ENSDART00000134677 ENSDART00000130800 |
cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr22_+_2709467 | 1.73 |
ENSDART00000141416
|
znf1171
|
zinc finger protein 1171 |
| chr4_-_64482414 | 1.73 |
ENSDART00000159000
|
znf1105
|
zinc finger protein 1105 |
| chr10_-_11353101 | 1.72 |
ENSDART00000092110
|
lin54
|
lin-54 DREAM MuvB core complex component |
| chr13_-_23611960 | 1.72 |
ENSDART00000146216
|
prim2
|
DNA primase subunit 2 |
| chr7_+_38808027 | 1.72 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr22_+_13886821 | 1.71 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr3_-_16413606 | 1.70 |
ENSDART00000127309
ENSDART00000017172 ENSDART00000136465 |
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr23_+_16928506 | 1.70 |
ENSDART00000162292
ENSDART00000080625 |
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr5_+_12836913 | 1.69 |
ENSDART00000023101
|
pes
|
pescadillo |
| chr13_+_51710725 | 1.68 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr21_+_17051478 | 1.68 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr4_-_42294516 | 1.68 |
ENSDART00000133558
|
si:dkey-4e4.1
|
si:dkey-4e4.1 |
| chr4_+_64577406 | 1.68 |
ENSDART00000159754
|
si:ch211-223a21.4
|
si:ch211-223a21.4 |
| chr4_+_33461796 | 1.67 |
ENSDART00000150445
|
si:dkey-247i3.1
|
si:dkey-247i3.1 |
| chr17_-_43428397 | 1.67 |
ENSDART00000043866
|
plk4
|
polo-like kinase 4 (Drosophila) |
| chr4_-_22363709 | 1.67 |
ENSDART00000037670
|
orc5
|
origin recognition complex, subunit 5 |
| chr10_-_23099809 | 1.66 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr20_+_47143900 | 1.65 |
ENSDART00000153360
|
si:dkeyp-104f11.6
|
si:dkeyp-104f11.6 |
| chr4_-_19693978 | 1.64 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr19_+_28256076 | 1.64 |
ENSDART00000133354
|
irx4b
|
iroquois homeobox 4b |
| chr14_-_25111496 | 1.64 |
ENSDART00000158108
|
si:rp71-1d10.8
|
si:rp71-1d10.8 |
| chr5_-_29531948 | 1.63 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr16_+_48460873 | 1.63 |
ENSDART00000159902
|
ext1a
|
exostosin glycosyltransferase 1a |
| chr16_-_43233509 | 1.63 |
ENSDART00000025877
|
cldn12
|
claudin 12 |
| chr22_-_36829006 | 1.63 |
ENSDART00000170256
ENSDART00000086064 |
map1sa
|
microtubule-associated protein 1Sa |
| chr13_-_11984867 | 1.62 |
ENSDART00000157538
|
npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr4_-_12477224 | 1.61 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr2_+_10821127 | 1.61 |
ENSDART00000145770
ENSDART00000174629 ENSDART00000081094 |
glmna
|
glomulin, FKBP associated protein a |
| chr4_-_45301719 | 1.61 |
ENSDART00000150282
|
si:ch211-162i8.2
|
si:ch211-162i8.2 |
| chr16_-_12060488 | 1.61 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr15_+_37545855 | 1.61 |
ENSDART00000099456
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr7_+_70338270 | 1.58 |
ENSDART00000065234
|
gba3
|
glucosidase, beta, acid 3 (gene/pseudogene) |
| chr3_-_40162843 | 1.57 |
ENSDART00000129664
ENSDART00000025285 |
drg2
|
developmentally regulated GTP binding protein 2 |
| chr11_-_3907937 | 1.56 |
ENSDART00000082409
|
rpn1
|
ribophorin I |
| chr23_+_25708787 | 1.55 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr15_-_41689684 | 1.55 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr23_-_24146591 | 1.55 |
ENSDART00000133269
|
arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr17_+_17764979 | 1.54 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr8_-_47206114 | 1.54 |
ENSDART00000147606
ENSDART00000060883 ENSDART00000060880 |
ppih
|
peptidylprolyl isomerase H (cyclophilin H) |
| chr16_-_5115993 | 1.53 |
ENSDART00000138654
|
ttk
|
ttk protein kinase |
| chr16_+_31942527 | 1.53 |
ENSDART00000188334
ENSDART00000188643 ENSDART00000058496 |
phc1
|
polyhomeotic homolog 1 |
| chr11_-_21218172 | 1.53 |
ENSDART00000027532
|
mapkapk2a
|
mitogen-activated protein kinase-activated protein kinase 2a |
| chr3_-_15734358 | 1.53 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr17_-_22573311 | 1.53 |
ENSDART00000141523
ENSDART00000140022 ENSDART00000079390 ENSDART00000188644 |
exo1
|
exonuclease 1 |
| chr19_+_3213914 | 1.53 |
ENSDART00000193144
|
zgc:86598
|
zgc:86598 |
| chr4_-_50544015 | 1.52 |
ENSDART00000155155
|
zgc:174704
|
zgc:174704 |
| chr10_-_28380919 | 1.52 |
ENSDART00000183409
ENSDART00000183105 ENSDART00000100207 ENSDART00000185392 ENSDART00000131220 |
btg3
|
B-cell translocation gene 3 |
| chr14_-_41468892 | 1.51 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr15_+_1796313 | 1.50 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr23_+_34990693 | 1.49 |
ENSDART00000013449
|
CHST13
|
si:ch211-236h17.3 |
| chr11_-_6970107 | 1.49 |
ENSDART00000171255
|
COMP
|
si:ch211-43f4.1 |
| chr17_-_7792376 | 1.49 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr6_+_43426599 | 1.49 |
ENSDART00000056457
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr12_+_19199735 | 1.49 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr7_-_24364536 | 1.48 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr16_-_54942532 | 1.48 |
ENSDART00000078887
ENSDART00000101402 |
tmem222a
|
transmembrane protein 222a |
| chr11_-_22372072 | 1.48 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr17_+_37932433 | 1.48 |
ENSDART00000185349
|
plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr15_+_823437 | 1.48 |
ENSDART00000155608
|
si:dkey-7i4.7
|
si:dkey-7i4.7 |
| chr2_+_10821579 | 1.47 |
ENSDART00000179694
|
glmna
|
glomulin, FKBP associated protein a |
| chr3_-_49514874 | 1.47 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr22_-_18746508 | 1.46 |
ENSDART00000003929
|
midn
|
midnolin |
| chr23_+_20513104 | 1.46 |
ENSDART00000079591
|
dpm1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr5_-_67365750 | 1.46 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr7_+_46019780 | 1.46 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr15_+_28685892 | 1.45 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr4_-_56898328 | 1.45 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr4_-_69615167 | 1.45 |
ENSDART00000171108
|
si:ch211-120c15.3
|
si:ch211-120c15.3 |
| chr17_+_956821 | 1.45 |
ENSDART00000082101
|
ppp2r5ca
|
protein phosphatase 2, regulatory subunit B', gamma a |
| chr20_+_39223235 | 1.44 |
ENSDART00000132132
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr4_+_30454496 | 1.44 |
ENSDART00000164555
|
si:dkey-199m13.4
|
si:dkey-199m13.4 |
| chr25_-_32751982 | 1.44 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr23_-_3408777 | 1.43 |
ENSDART00000193245
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr4_-_36144500 | 1.42 |
ENSDART00000170896
|
znf992
|
zinc finger protein 992 |
| chr2_-_17115256 | 1.41 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr4_+_332709 | 1.41 |
ENSDART00000067486
|
tulp4a
|
tubby like protein 4a |
| chr19_+_14454306 | 1.40 |
ENSDART00000161965
|
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr4_-_26035770 | 1.40 |
ENSDART00000124514
|
usp44
|
ubiquitin specific peptidase 44 |
| chr18_-_50524017 | 1.40 |
ENSDART00000150013
ENSDART00000149912 |
cd276
|
CD276 molecule |
| chr9_-_31524907 | 1.39 |
ENSDART00000142904
ENSDART00000127214 ENSDART00000133427 ENSDART00000146268 ENSDART00000182541 ENSDART00000184736 |
tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr8_+_20863025 | 1.39 |
ENSDART00000015891
ENSDART00000187089 ENSDART00000146989 |
fzr1a
|
fizzy/cell division cycle 20 related 1a |
| chr17_-_24866727 | 1.39 |
ENSDART00000027957
|
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr20_-_51559419 | 1.39 |
ENSDART00000065231
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr18_+_14633974 | 1.39 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr10_-_11353469 | 1.38 |
ENSDART00000191847
|
lin54
|
lin-54 DREAM MuvB core complex component |
| chr3_+_13879446 | 1.38 |
ENSDART00000164767
|
farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr16_+_54209504 | 1.37 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr6_-_11614339 | 1.37 |
ENSDART00000080589
|
gulp1b
|
GULP, engulfment adaptor PTB domain containing 1b |
| chr18_-_8380090 | 1.37 |
ENSDART00000141581
ENSDART00000081143 |
sephs1
|
selenophosphate synthetase 1 |
| chr4_+_77141970 | 1.37 |
ENSDART00000174209
|
si:dkey-172k15.11
|
si:dkey-172k15.11 |
| chr4_-_77218637 | 1.37 |
ENSDART00000174325
|
psmb10
|
proteasome subunit beta 10 |
| chr20_-_3319642 | 1.37 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr16_-_4769877 | 1.36 |
ENSDART00000149421
ENSDART00000054078 |
rpa2
|
replication protein A2 |
| chr4_+_58576146 | 1.36 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr17_-_9995667 | 1.36 |
ENSDART00000148463
|
snx6
|
sorting nexin 6 |
| chr12_-_13205854 | 1.36 |
ENSDART00000077829
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr23_-_20126257 | 1.36 |
ENSDART00000005021
|
tktb
|
transketolase b |
| chr23_-_43424510 | 1.35 |
ENSDART00000055564
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr1_-_36772147 | 1.35 |
ENSDART00000053369
|
prmt9
|
protein arginine methyltransferase 9 |
| chr14_+_35369979 | 1.35 |
ENSDART00000144702
ENSDART00000169712 |
clint1a
|
clathrin interactor 1a |
| chr15_-_25584888 | 1.34 |
ENSDART00000127571
|
si:dkey-54n8.2
|
si:dkey-54n8.2 |
| chr16_-_4770233 | 1.34 |
ENSDART00000193228
|
rpa2
|
replication protein A2 |
| chr21_-_4849029 | 1.33 |
ENSDART00000168930
ENSDART00000151019 |
notch1a
|
notch 1a |
| chr19_-_29303788 | 1.32 |
ENSDART00000112167
|
srfbp1
|
serum response factor binding protein 1 |
| chr3_+_14641962 | 1.32 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr21_+_18907102 | 1.31 |
ENSDART00000160185
ENSDART00000190175 ENSDART00000017937 ENSDART00000191546 ENSDART00000130519 ENSDART00000137143 |
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr12_-_27212596 | 1.31 |
ENSDART00000153101
|
psme3
|
proteasome activator subunit 3 |
| chr2_-_43739559 | 1.31 |
ENSDART00000138947
|
kif5ba
|
kinesin family member 5B, a |
| chr13_+_502230 | 1.30 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr8_+_12951155 | 1.30 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr15_-_41689981 | 1.30 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr11_+_45388126 | 1.30 |
ENSDART00000167826
ENSDART00000171274 |
srsf11
|
serine/arginine-rich splicing factor 11 |
| chr9_-_28939181 | 1.29 |
ENSDART00000101276
ENSDART00000135334 |
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr20_-_54245256 | 1.29 |
ENSDART00000170482
|
prpf39
|
PRP39 pre-mRNA processing factor 39 homolog (yeast) |
Network of associatons between targets according to the STRING database.
First level regulatory network of atf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 1.0 | 3.9 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.9 | 3.7 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.8 | 3.4 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.8 | 3.1 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.8 | 2.3 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.7 | 3.7 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.7 | 9.5 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.7 | 3.6 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.7 | 2.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.7 | 2.0 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.6 | 2.5 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.6 | 1.9 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.6 | 1.8 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.5 | 2.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.5 | 2.1 | GO:0051661 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.5 | 2.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.5 | 0.9 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.5 | 1.4 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.5 | 1.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 2.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.4 | 1.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.4 | 2.5 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.4 | 1.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.4 | 2.0 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.4 | 1.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.4 | 1.9 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.4 | 1.5 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 1.1 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.3 | 1.0 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.3 | 1.0 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.3 | 1.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.3 | 2.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.3 | 1.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.3 | 0.9 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) negative regulation of cell maturation(GO:1903430) |
| 0.3 | 1.5 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.3 | 1.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.3 | 1.5 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.3 | 2.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 0.9 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 1.5 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.3 | 1.5 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.3 | 1.2 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.3 | 2.0 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.3 | 0.8 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.3 | 1.4 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.3 | 0.8 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.3 | 2.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.3 | 2.9 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.3 | 1.3 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.3 | 1.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.2 | 0.7 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.2 | 1.4 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 1.4 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.2 | 2.1 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.2 | 1.4 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.2 | 1.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 1.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 0.7 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 2.7 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.2 | 3.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 1.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 2.3 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.2 | 1.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.2 | 1.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 2.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 0.8 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.2 | 0.6 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.2 | 1.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.2 | 3.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 2.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 1.7 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 0.8 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 0.5 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.2 | 0.8 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.2 | 3.0 | GO:1902806 | regulation of cell cycle G1/S phase transition(GO:1902806) |
| 0.2 | 0.5 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.2 | 2.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 1.3 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.2 | 1.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 1.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 1.1 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.2 | 1.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 0.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 1.4 | GO:0042987 | Notch receptor processing(GO:0007220) beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 4.3 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.1 | 3.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 1.2 | GO:0072502 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 1.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 1.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.1 | 0.8 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 1.4 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 1.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 1.8 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) heterochromatin organization(GO:0070828) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.4 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 1.2 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.1 | 2.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 1.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.6 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 0.4 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.9 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 1.1 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.1 | 2.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 1.0 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 1.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.7 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.1 | 1.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.6 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.5 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.6 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.8 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 1.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.8 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 1.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 2.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.4 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 1.1 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 0.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.9 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.6 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 1.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 2.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 1.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 2.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.9 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.5 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.4 | GO:0071632 | optomotor response(GO:0071632) |
| 0.1 | 4.2 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 2.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 1.8 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.1 | 1.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 2.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.3 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.1 | 0.8 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.9 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 2.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 2.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.2 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 0.7 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 1.0 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 1.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 1.8 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 1.4 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 1.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 2.1 | GO:0007379 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.1 | 1.0 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.7 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 1.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.5 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 1.5 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.1 | 0.6 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 5.4 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.6 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 1.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.8 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 2.7 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.1 | 0.5 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 0.4 | GO:0003343 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.1 | 0.3 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.1 | 0.8 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.3 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.1 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.3 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 1.6 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.8 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.8 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 1.1 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.1 | 0.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 2.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.0 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 2.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 5.9 | GO:0032880 | regulation of protein localization(GO:0032880) |
| 0.1 | 0.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 5.2 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 1.6 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 1.5 | GO:0014020 | neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) |
| 0.1 | 0.7 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 1.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 1.4 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.4 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.8 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.1 | 0.5 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 2.1 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.1 | 1.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.7 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 1.6 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 0.5 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.1 | 0.7 | GO:0034033 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.3 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 1.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.8 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 1.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 2.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.2 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.4 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 1.5 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 1.9 | GO:0030335 | positive regulation of cell migration(GO:0030335) |
| 0.0 | 2.4 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.7 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.6 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.8 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.0 | 0.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.8 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.5 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.9 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 3.0 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.8 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.7 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 1.6 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 2.5 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.3 | GO:0051340 | regulation of ligase activity(GO:0051340) positive regulation of ligase activity(GO:0051351) |
| 0.0 | 0.7 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.5 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 0.5 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 1.4 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.9 | GO:0051961 | negative regulation of neurogenesis(GO:0050768) negative regulation of nervous system development(GO:0051961) |
| 0.0 | 0.4 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 1.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.5 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.5 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 1.5 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.3 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 79.9 | GO:0006355 | regulation of transcription, DNA-templated(GO:0006355) |
| 0.0 | 2.4 | GO:0061371 | determination of heart left/right asymmetry(GO:0061371) |
| 0.0 | 1.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 1.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 4.5 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 1.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.5 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.4 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 1.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 1.5 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.5 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 1.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 2.7 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 0.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.0 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:1990077 | primosome complex(GO:1990077) |
| 1.2 | 3.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.8 | 5.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.6 | 2.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.6 | 3.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.6 | 2.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.6 | 1.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.5 | 3.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.5 | 7.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.4 | 2.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 1.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 1.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.4 | 2.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 2.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 2.9 | GO:0035101 | FACT complex(GO:0035101) |
| 0.3 | 2.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.3 | 1.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.3 | 1.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 1.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.3 | 2.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 0.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 0.9 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 1.6 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 1.8 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.2 | 1.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.2 | 1.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 2.6 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.2 | 2.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 1.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.2 | 0.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 0.9 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.2 | 2.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 3.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 2.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 1.8 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.9 | GO:0016234 | inclusion body(GO:0016234) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.0 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.5 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 3.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 2.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 2.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 2.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 3.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 2.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.8 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 5.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 1.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.4 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 4.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.9 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 3.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 29.2 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 4.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 5.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 4.6 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 1.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 98.9 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.2 | GO:0019867 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 1.0 | 3.1 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.9 | 2.6 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.8 | 2.4 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.8 | 2.3 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.6 | 1.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.6 | 1.7 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.5 | 1.5 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.5 | 2.0 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.5 | 1.5 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.4 | 1.8 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.4 | 2.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.4 | 2.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 1.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.4 | 1.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) efflux transmembrane transporter activity(GO:0015562) |
| 0.4 | 1.5 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.4 | 1.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.4 | 1.1 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 1.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 4.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.3 | 1.0 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.3 | 1.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.3 | 1.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.3 | 1.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.3 | 1.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.3 | 1.4 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.3 | 1.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.3 | 1.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 4.1 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 1.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 1.4 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.2 | 2.0 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.2 | 1.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 1.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.2 | 1.8 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 4.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 0.9 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.2 | 1.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 0.5 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.2 | 1.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 1.4 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 2.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 1.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 0.5 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.2 | 0.6 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 0.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 12.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 5.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.6 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 1.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.6 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 5.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 1.0 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.5 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 1.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 1.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.3 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.0 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 1.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 2.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.1 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 1.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.0 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 2.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 3.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 1.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.4 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 1.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.6 | GO:0051219 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.1 | 0.8 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.3 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.1 | 1.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.2 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.9 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 2.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 1.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 2.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.4 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 2.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 2.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 4.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.5 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.2 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0070717 | poly(A) binding(GO:0008143) poly-purine tract binding(GO:0070717) |
| 0.0 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 3.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 3.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 92.1 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 3.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.7 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 6.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.6 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.2 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 3.3 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 3.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 2.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 8.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 3.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 2.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 3.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.4 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 1.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.4 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.9 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 6.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 5.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.9 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 1.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 1.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 1.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.8 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.5 | 2.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 6.4 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.3 | 2.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 1.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 5.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 3.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 1.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 2.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 0.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 2.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 4.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 2.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 3.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.4 | REACTOME BASE EXCISION REPAIR | Genes involved in Base Excision Repair |
| 0.1 | 2.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 3.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 4.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 2.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 1.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.8 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.6 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |