Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
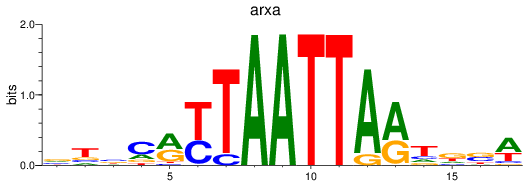
Results for arxa
Z-value: 0.52
Transcription factors associated with arxa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arxa
|
ENSDARG00000058011 | aristaless related homeobox a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arxa | dr11_v1_chr24_-_23942722_23942722 | -0.18 | 4.6e-01 | Click! |
Activity profile of arxa motif
Sorted Z-values of arxa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_22320738 | 1.31 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr3_-_19368435 | 1.31 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr12_-_35830625 | 1.27 |
ENSDART00000180028
|
CU459056.1
|
|
| chr25_-_21031007 | 1.22 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr2_-_26596794 | 1.18 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr16_+_54209504 | 1.13 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr20_+_11731039 | 1.10 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr8_-_39822917 | 1.09 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr23_-_42232124 | 1.07 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr3_-_26017831 | 1.07 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr8_-_50888806 | 0.98 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr2_-_38284648 | 0.98 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr23_-_16485190 | 0.97 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr24_+_19415124 | 0.94 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr20_+_29209615 | 0.92 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr25_+_30131055 | 0.91 |
ENSDART00000152705
|
api5
|
apoptosis inhibitor 5 |
| chr9_-_34937025 | 0.90 |
ENSDART00000137888
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr24_-_38657683 | 0.88 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr9_-_1986014 | 0.87 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr8_+_26059677 | 0.85 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr23_+_1029450 | 0.85 |
ENSDART00000189196
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr23_+_12134839 | 0.84 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr17_-_9950911 | 0.81 |
ENSDART00000105117
|
sptssa
|
serine palmitoyltransferase, small subunit A |
| chr25_+_20715950 | 0.80 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr7_+_2969661 | 0.79 |
ENSDART00000158382
|
CABZ01007829.1
|
|
| chr13_-_12602920 | 0.78 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr2_-_9989919 | 0.77 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr6_-_55399214 | 0.77 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr16_-_7793457 | 0.77 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr21_+_43172506 | 0.74 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr17_-_16422654 | 0.73 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr4_+_11723852 | 0.72 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr16_-_27677930 | 0.71 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr19_-_5669122 | 0.71 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr22_-_36530902 | 0.70 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr22_-_5252005 | 0.70 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr11_-_21528056 | 0.69 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr25_+_11008419 | 0.68 |
ENSDART00000156589
|
mhc1lia
|
major histocompatibility complex class I LIA |
| chr7_-_8712148 | 0.68 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr8_+_17167876 | 0.67 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr3_+_30501135 | 0.67 |
ENSDART00000165869
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr11_-_39118882 | 0.65 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr16_+_23799622 | 0.65 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr8_+_28259347 | 0.65 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr11_-_41853874 | 0.64 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr6_+_7533601 | 0.63 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr15_+_23799461 | 0.61 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr14_+_34558480 | 0.61 |
ENSDART00000075170
|
pttg1
|
pituitary tumor-transforming 1 |
| chr10_-_2788668 | 0.61 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr12_-_28363111 | 0.60 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr8_+_49065348 | 0.60 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr2_-_10877765 | 0.60 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr5_-_54672763 | 0.59 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr8_+_25079470 | 0.59 |
ENSDART00000000744
|
sypl2b
|
synaptophysin-like 2b |
| chr18_-_46258612 | 0.59 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr20_+_29209767 | 0.59 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr15_-_4528326 | 0.58 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr2_+_10878406 | 0.57 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr1_+_21731382 | 0.56 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr13_-_31017960 | 0.56 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr14_-_41388178 | 0.56 |
ENSDART00000124532
ENSDART00000125016 ENSDART00000169247 |
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr23_+_36460239 | 0.55 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr5_-_69649044 | 0.54 |
ENSDART00000011192
ENSDART00000170885 |
eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha |
| chr3_+_40164129 | 0.54 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr7_-_5375214 | 0.54 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr4_-_9909371 | 0.53 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr4_+_19700308 | 0.52 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr21_+_6394929 | 0.52 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr18_+_39937225 | 0.52 |
ENSDART00000141136
|
si:ch211-282k23.2
|
si:ch211-282k23.2 |
| chr2_-_45331115 | 0.51 |
ENSDART00000158003
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr23_-_36303216 | 0.51 |
ENSDART00000188720
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr20_+_25225112 | 0.51 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr2_+_9990491 | 0.51 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr1_+_27977297 | 0.50 |
ENSDART00000180692
ENSDART00000166819 |
sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr2_+_2169337 | 0.50 |
ENSDART00000179939
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr11_+_31324335 | 0.50 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr6_+_36839509 | 0.50 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr20_+_25586099 | 0.50 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr7_-_60156409 | 0.49 |
ENSDART00000006802
|
cct7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr17_-_29224908 | 0.49 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr1_-_45616470 | 0.49 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr21_-_15200556 | 0.48 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr9_-_11676491 | 0.48 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr10_+_26652859 | 0.48 |
ENSDART00000079174
|
htatsf1
|
HIV-1 Tat specific factor 1 |
| chr14_+_5385855 | 0.47 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr25_+_7241084 | 0.47 |
ENSDART00000190501
ENSDART00000190588 |
hmg20a
|
high mobility group 20A |
| chr20_+_29209926 | 0.47 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr8_+_18011522 | 0.47 |
ENSDART00000136756
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr25_-_28674739 | 0.46 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr19_+_2631565 | 0.46 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr16_-_30434279 | 0.46 |
ENSDART00000018504
|
zgc:77086
|
zgc:77086 |
| chr21_+_43328685 | 0.45 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr10_+_42521943 | 0.45 |
ENSDART00000010420
ENSDART00000075303 |
actr1
|
ARP1 actin related protein 1, centractin |
| chr6_-_40922971 | 0.44 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr14_-_5817039 | 0.43 |
ENSDART00000131820
|
kazald2
|
Kazal-type serine peptidase inhibitor domain 2 |
| chr11_+_16153207 | 0.43 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr12_-_36521767 | 0.42 |
ENSDART00000110290
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr6_+_40922572 | 0.42 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr23_+_4709607 | 0.42 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr23_-_33350990 | 0.42 |
ENSDART00000144831
|
si:ch211-226m16.2
|
si:ch211-226m16.2 |
| chr7_+_24023653 | 0.42 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr19_+_42955280 | 0.42 |
ENSDART00000141225
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr10_-_3138859 | 0.41 |
ENSDART00000190606
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr10_-_3138403 | 0.40 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr18_+_39106722 | 0.40 |
ENSDART00000122377
|
bcl2l10
|
BCL2 like 10 |
| chr7_-_51368681 | 0.40 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr6_+_25261297 | 0.40 |
ENSDART00000162824
ENSDART00000163490 ENSDART00000157790 ENSDART00000160978 ENSDART00000161545 ENSDART00000159978 |
kyat3
|
kynurenine aminotransferase 3 |
| chr23_+_29885019 | 0.40 |
ENSDART00000167059
|
aurkaip1
|
aurora kinase A interacting protein 1 |
| chr5_+_32076109 | 0.39 |
ENSDART00000051357
ENSDART00000144510 |
zmat5
|
zinc finger, matrin-type 5 |
| chr7_+_19495905 | 0.39 |
ENSDART00000125584
ENSDART00000173774 |
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr6_-_7720332 | 0.38 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr15_-_35098345 | 0.37 |
ENSDART00000181824
|
CR387932.1
|
|
| chr11_-_16152400 | 0.37 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr12_-_48188928 | 0.37 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr5_+_66433287 | 0.36 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr5_-_52010122 | 0.35 |
ENSDART00000073627
ENSDART00000163898 ENSDART00000051003 |
cdk7
|
cyclin-dependent kinase 7 |
| chr11_+_31323746 | 0.35 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr14_+_30568961 | 0.35 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr9_-_30555725 | 0.35 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr24_+_38301080 | 0.34 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr7_-_30677104 | 0.34 |
ENSDART00000188360
|
myo1ea
|
myosin IE, a |
| chr24_+_29912509 | 0.34 |
ENSDART00000168422
|
frrs1b
|
ferric-chelate reductase 1b |
| chr11_+_16152316 | 0.33 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr13_-_35808904 | 0.33 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr8_-_53044300 | 0.32 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr17_-_10122204 | 0.32 |
ENSDART00000160751
|
BX088587.1
|
|
| chr22_+_11775269 | 0.31 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr21_-_30082414 | 0.31 |
ENSDART00000157307
ENSDART00000155188 |
ccnjl
|
cyclin J-like |
| chr2_-_59205393 | 0.30 |
ENSDART00000056417
ENSDART00000182452 ENSDART00000141876 |
ftr30
|
finTRIM family, member 30 |
| chr4_-_1801519 | 0.30 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr3_-_34052882 | 0.30 |
ENSDART00000151463
|
ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr9_-_41062412 | 0.30 |
ENSDART00000193879
|
ankar
|
ankyrin and armadillo repeat containing |
| chr3_+_27798094 | 0.30 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr9_+_43799829 | 0.30 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr3_-_34034498 | 0.29 |
ENSDART00000151145
|
ighv11-2
|
immunoglobulin heavy variable 11-2 |
| chr1_+_47499888 | 0.28 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr18_+_7073130 | 0.28 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr7_-_50410524 | 0.28 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr3_-_26787430 | 0.28 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr13_-_4707018 | 0.27 |
ENSDART00000128422
|
oit3
|
oncoprotein induced transcript 3 |
| chr17_+_20589553 | 0.26 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr2_-_17392799 | 0.26 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr7_+_21272833 | 0.25 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr23_-_30041065 | 0.25 |
ENSDART00000131209
ENSDART00000127192 |
ccdc187
|
coiled-coil domain containing 187 |
| chr19_-_10810006 | 0.25 |
ENSDART00000151157
|
si:dkey-3n22.9
|
si:dkey-3n22.9 |
| chr20_-_37813863 | 0.25 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr16_+_23984179 | 0.24 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr12_-_4185554 | 0.24 |
ENSDART00000152313
|
si:dkey-32n7.7
|
si:dkey-32n7.7 |
| chr20_-_51355465 | 0.24 |
ENSDART00000151620
ENSDART00000151690 ENSDART00000110289 |
tcte1
|
t-complex-associated-testis-expressed 1 |
| chr11_-_1509773 | 0.24 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr5_+_40485503 | 0.24 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr8_+_18615938 | 0.24 |
ENSDART00000089161
|
si:ch211-232d19.4
|
si:ch211-232d19.4 |
| chr20_+_40457599 | 0.23 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr13_-_36798204 | 0.23 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr21_+_13387965 | 0.23 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr19_+_43780970 | 0.22 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr10_+_38610741 | 0.22 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr1_-_19502322 | 0.22 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr2_+_11923615 | 0.22 |
ENSDART00000126118
|
trove2
|
TROVE domain family, member 2 |
| chr23_-_31969786 | 0.21 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr10_-_43771447 | 0.21 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr21_-_4793686 | 0.21 |
ENSDART00000158232
|
notch1a
|
notch 1a |
| chr4_-_45115963 | 0.20 |
ENSDART00000137248
|
si:dkey-51d8.6
|
si:dkey-51d8.6 |
| chr24_+_35787629 | 0.20 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr6_+_12527725 | 0.20 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr6_+_49551614 | 0.20 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr21_+_45223194 | 0.19 |
ENSDART00000150902
|
si:ch73-269m14.3
|
si:ch73-269m14.3 |
| chr5_-_11809710 | 0.19 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr2_+_1988036 | 0.19 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr24_+_37640626 | 0.19 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr9_+_22351443 | 0.19 |
ENSDART00000080054
|
crygs3
|
crystallin, gamma S3 |
| chr5_+_24156170 | 0.19 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr5_+_33498253 | 0.19 |
ENSDART00000140993
|
ms4a17c.2
|
membrane-spanning 4-domains, subfamily A, member 17c.2 |
| chr21_-_45363871 | 0.18 |
ENSDART00000075443
ENSDART00000182078 ENSDART00000151106 |
PPP2CA
|
zgc:56064 |
| chr7_+_26173751 | 0.17 |
ENSDART00000065131
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
| chr6_+_8598428 | 0.17 |
ENSDART00000032118
|
ube2g2
|
ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) |
| chr20_-_51831816 | 0.17 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr4_-_69428110 | 0.17 |
ENSDART00000160607
|
si:ch211-145h19.3
|
si:ch211-145h19.3 |
| chr24_-_37640705 | 0.17 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr7_+_19495379 | 0.17 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr20_+_38458084 | 0.16 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr8_-_51930826 | 0.16 |
ENSDART00000109785
|
cabin1
|
calcineurin binding protein 1 |
| chr7_+_22543963 | 0.16 |
ENSDART00000101528
|
chrnb1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr14_-_470505 | 0.15 |
ENSDART00000067147
|
ANKRD50
|
ankyrin repeat domain 50 |
| chr16_+_13818743 | 0.15 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr10_+_34010494 | 0.15 |
ENSDART00000186836
ENSDART00000114703 |
kl
|
klotho |
| chr20_+_22799857 | 0.15 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr21_-_19314618 | 0.15 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr7_+_34453185 | 0.15 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr3_+_32118670 | 0.15 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr16_+_35379720 | 0.15 |
ENSDART00000170497
|
si:dkey-34d22.2
|
si:dkey-34d22.2 |
| chr2_-_17393216 | 0.14 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr4_-_5108844 | 0.14 |
ENSDART00000132666
ENSDART00000136096 |
tmem209
|
transmembrane protein 209 |
| chr18_+_17827149 | 0.14 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr20_+_20731633 | 0.14 |
ENSDART00000191952
ENSDART00000165224 |
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr13_+_7049823 | 0.14 |
ENSDART00000178997
ENSDART00000161443 |
rnaset2
|
ribonuclease T2 |
| chr3_+_32365811 | 0.14 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr15_-_21014270 | 0.13 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr2_+_35806672 | 0.13 |
ENSDART00000137384
|
rasal2
|
RAS protein activator like 2 |
| chr22_+_737211 | 0.13 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
Network of associatons between targets according to the STRING database.
First level regulatory network of arxa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.4 | 1.1 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.3 | 1.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 0.5 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 1.1 | GO:0097065 | anterior head development(GO:0097065) |
| 0.2 | 0.7 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 0.5 | GO:0046333 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.2 | 0.8 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.7 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.7 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.9 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.5 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.1 | 0.7 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.9 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.5 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.9 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.2 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.1 | 0.6 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 0.2 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.6 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.2 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0051279 | regulation of calcium ion transport into cytosol(GO:0010522) regulation of release of sequestered calcium ion into cytosol(GO:0051279) |
| 0.0 | 1.0 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.5 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.5 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.5 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.4 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.6 | GO:0033046 | negative regulation of sister chromatid segregation(GO:0033046) negative regulation of mitotic sister chromatid segregation(GO:0033048) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.8 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.0 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 1.0 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.7 | GO:0045335 | dendritic spine head(GO:0044327) phagocytic vesicle(GO:0045335) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 0.7 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 0.5 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 0.5 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.2 | 1.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 0.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.8 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.3 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.8 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.4 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 1.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.7 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |