Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
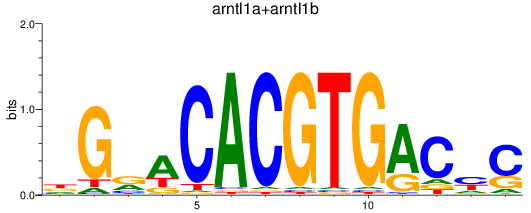
Results for arntl1a+arntl1b
Z-value: 0.65
Transcription factors associated with arntl1a+arntl1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arntl1a
|
ENSDARG00000006791 | aryl hydrocarbon receptor nuclear translocator-like 1a |
|
arntl1b
|
ENSDARG00000035732 | aryl hydrocarbon receptor nuclear translocator-like 1b |
|
arntl1b
|
ENSDARG00000114562 | aryl hydrocarbon receptor nuclear translocator-like 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arntl1a | dr11_v1_chr25_-_17910714_17910714 | -0.64 | 3.2e-03 | Click! |
| arntl1b | dr11_v1_chr7_+_66048102_66048102 | 0.53 | 2.1e-02 | Click! |
Activity profile of arntl1a+arntl1b motif
Sorted Z-values of arntl1a+arntl1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_13696537 | 2.93 |
ENSDART00000109195
ENSDART00000122041 ENSDART00000192554 |
nrcama
|
neuronal cell adhesion molecule a |
| chr7_+_48999723 | 2.65 |
ENSDART00000182699
ENSDART00000166329 |
si:ch211-288d18.1
|
si:ch211-288d18.1 |
| chr13_-_293250 | 2.35 |
ENSDART00000138581
|
chs1
|
chitin synthase 1 |
| chr9_+_42066030 | 1.90 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr1_-_54765262 | 1.62 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr13_-_3474373 | 1.59 |
ENSDART00000157437
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr15_-_34418525 | 1.46 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr21_+_43178831 | 1.30 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr21_-_4539899 | 1.26 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr25_-_35599887 | 1.21 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr18_+_26337869 | 1.21 |
ENSDART00000109257
|
RASGRF1
|
si:ch211-234p18.3 |
| chr24_+_3307857 | 1.19 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr9_+_42095220 | 1.17 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr16_+_26824691 | 1.12 |
ENSDART00000135053
|
zmp:0000001316
|
zmp:0000001316 |
| chr8_-_25235676 | 1.10 |
ENSDART00000062363
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr6_-_39605734 | 1.06 |
ENSDART00000044276
ENSDART00000179059 |
dip2bb
|
disco-interacting protein 2 homolog Bb |
| chr3_-_40955780 | 1.03 |
ENSDART00000130130
|
cyp3c3
|
cytochrome P450, family 3, subfamily c, polypeptide 3 |
| chr20_-_2949028 | 1.02 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr17_+_33340675 | 1.00 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr15_-_37850969 | 0.99 |
ENSDART00000031418
|
hsc70
|
heat shock cognate 70 |
| chr13_+_19884631 | 0.99 |
ENSDART00000089533
|
atrnl1a
|
attractin-like 1a |
| chr6_-_49547680 | 0.98 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr17_+_30448452 | 0.98 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr22_-_94352 | 0.97 |
ENSDART00000184883
|
ndufaf3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr21_-_41588129 | 0.96 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr20_+_40766645 | 0.92 |
ENSDART00000144401
|
tbc1d32
|
TBC1 domain family, member 32 |
| chr1_+_6189949 | 0.91 |
ENSDART00000092277
|
retreg2
|
reticulophagy regulator family member 2 |
| chr23_-_16692312 | 0.89 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr11_-_43104475 | 0.86 |
ENSDART00000125368
|
acyp2
|
acylphosphatase 2, muscle type |
| chr24_-_7632187 | 0.86 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr10_-_44027391 | 0.86 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr9_-_1200187 | 0.85 |
ENSDART00000158760
|
ino80da
|
INO80 complex subunit Da |
| chr19_+_40379771 | 0.84 |
ENSDART00000017917
ENSDART00000110699 |
vps50
vps50
|
VPS50 EARP/GARPII complex subunit VPS50 EARP/GARPII complex subunit |
| chr11_+_44300548 | 0.83 |
ENSDART00000191626
|
CABZ01100304.1
|
|
| chr15_+_1199407 | 0.83 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr24_-_14711597 | 0.83 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr13_+_46803979 | 0.81 |
ENSDART00000159260
|
CU695232.1
|
|
| chr7_+_47243564 | 0.79 |
ENSDART00000098942
ENSDART00000162237 |
znf507
|
zinc finger protein 507 |
| chr1_+_7956030 | 0.78 |
ENSDART00000159655
|
CR855320.1
|
|
| chr18_+_2189211 | 0.77 |
ENSDART00000170827
|
ccpg1
|
cell cycle progression 1 |
| chr11_+_29770966 | 0.76 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr23_+_42131509 | 0.76 |
ENSDART00000184544
|
SHISAL2A
|
shisa like 2A |
| chr24_+_7631797 | 0.75 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr10_-_6775271 | 0.75 |
ENSDART00000110735
|
zgc:194281
|
zgc:194281 |
| chr21_+_31150438 | 0.74 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr2_+_6181383 | 0.73 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr2_+_33189582 | 0.73 |
ENSDART00000145588
ENSDART00000136330 ENSDART00000139295 ENSDART00000086340 |
rnf220a
|
ring finger protein 220a |
| chr21_-_42100471 | 0.73 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr25_+_34749187 | 0.72 |
ENSDART00000141473
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr5_-_10002260 | 0.70 |
ENSDART00000141831
|
si:ch73-266o15.4
|
si:ch73-266o15.4 |
| chr8_+_41533268 | 0.69 |
ENSDART00000142377
|
si:ch211-158d24.2
|
si:ch211-158d24.2 |
| chr25_+_37443194 | 0.69 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr10_-_41157135 | 0.67 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr8_+_46386601 | 0.67 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr22_+_38159823 | 0.66 |
ENSDART00000104527
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr2_+_4303125 | 0.65 |
ENSDART00000161211
ENSDART00000162269 |
cables1
|
Cdk5 and Abl enzyme substrate 1 |
| chr18_+_39074139 | 0.64 |
ENSDART00000142390
|
gnb5b
|
guanine nucleotide binding protein (G protein), beta 5b |
| chr15_-_43978141 | 0.63 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr9_-_3149896 | 0.63 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr9_-_18742704 | 0.61 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr1_+_9004719 | 0.61 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr15_+_1148074 | 0.60 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr5_+_15992655 | 0.60 |
ENSDART00000182148
|
znrf3
|
zinc and ring finger 3 |
| chr18_-_46280578 | 0.60 |
ENSDART00000131724
|
pld3
|
phospholipase D family, member 3 |
| chr15_-_44077937 | 0.59 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr7_+_1442059 | 0.59 |
ENSDART00000173391
|
si:cabz01090193.1
|
si:cabz01090193.1 |
| chr4_-_2867461 | 0.59 |
ENSDART00000160308
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr7_-_67214972 | 0.58 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr1_-_25648568 | 0.58 |
ENSDART00000135763
|
DCHS2
|
si:dkey-30k22.7 |
| chr4_-_31064105 | 0.58 |
ENSDART00000157670
|
si:dkey-11n14.1
|
si:dkey-11n14.1 |
| chr20_+_50115335 | 0.57 |
ENSDART00000031139
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr25_+_37268900 | 0.57 |
ENSDART00000156737
|
si:dkey-234i14.6
|
si:dkey-234i14.6 |
| chr1_+_56447107 | 0.56 |
ENSDART00000091924
|
CABZ01059392.1
|
|
| chr17_+_8183393 | 0.54 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr11_+_42478184 | 0.54 |
ENSDART00000089963
|
zgc:110286
|
zgc:110286 |
| chr16_-_9869056 | 0.54 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr1_+_50639416 | 0.53 |
ENSDART00000141977
|
herc3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr1_+_17527342 | 0.52 |
ENSDART00000139702
ENSDART00000140076 ENSDART00000005593 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr19_+_7152966 | 0.52 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr7_+_34620418 | 0.51 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr18_+_6479963 | 0.51 |
ENSDART00000092752
ENSDART00000136333 |
wash1
|
WAS protein family homolog 1 |
| chr7_+_8324506 | 0.51 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr20_-_1151265 | 0.50 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr21_-_3700334 | 0.50 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr24_-_13364311 | 0.50 |
ENSDART00000183808
|
KCNB2
|
si:dkey-192j17.1 |
| chr3_-_1190132 | 0.50 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr13_-_23666579 | 0.49 |
ENSDART00000192457
ENSDART00000179777 |
map3k21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr24_+_36339242 | 0.49 |
ENSDART00000105686
ENSDART00000142264 |
grnb
|
granulin b |
| chr4_-_2036620 | 0.49 |
ENSDART00000150490
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr15_-_41290415 | 0.49 |
ENSDART00000152157
|
si:dkey-75b17.1
|
si:dkey-75b17.1 |
| chr22_-_1237003 | 0.48 |
ENSDART00000169746
|
ampd2a
|
adenosine monophosphate deaminase 2a |
| chr20_-_33174899 | 0.48 |
ENSDART00000047834
|
nbas
|
neuroblastoma amplified sequence |
| chr20_+_13969414 | 0.48 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr19_+_34169055 | 0.47 |
ENSDART00000135592
ENSDART00000186043 |
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr4_+_331351 | 0.47 |
ENSDART00000132625
|
tulp4a
|
tubby like protein 4a |
| chr15_+_22394074 | 0.46 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr6_-_1553314 | 0.46 |
ENSDART00000077209
|
tpra1
|
transmembrane protein, adipocyte asscociated 1 |
| chr3_-_25420931 | 0.46 |
ENSDART00000109601
ENSDART00000182184 |
bptf
|
bromodomain PHD finger transcription factor |
| chr20_+_46213553 | 0.46 |
ENSDART00000100532
|
stx7l
|
syntaxin 7-like |
| chr1_+_45754868 | 0.45 |
ENSDART00000084512
|
pkn1a
|
protein kinase N1a |
| chr21_-_36948 | 0.44 |
ENSDART00000181230
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr15_-_1198886 | 0.44 |
ENSDART00000063285
|
lxn
|
latexin |
| chr15_-_1843831 | 0.43 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr2_+_24936766 | 0.43 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr18_+_31280984 | 0.43 |
ENSDART00000170285
ENSDART00000150608 ENSDART00000159720 |
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr16_+_19677484 | 0.43 |
ENSDART00000150588
|
tmem196b
|
transmembrane protein 196b |
| chr15_-_3094516 | 0.42 |
ENSDART00000179719
|
slitrk5a
|
SLIT and NTRK-like family, member 5a |
| chr14_+_2243 | 0.42 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr12_+_13652361 | 0.42 |
ENSDART00000182757
ENSDART00000152689 |
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr3_+_24511959 | 0.41 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr17_+_50074372 | 0.41 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr8_-_26961779 | 0.41 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr20_+_47491247 | 0.39 |
ENSDART00000113412
|
lin28b
|
lin-28 homolog B (C. elegans) |
| chr16_+_14216581 | 0.39 |
ENSDART00000113093
|
gba
|
glucosidase, beta, acid |
| chr24_-_42072886 | 0.39 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr7_+_7048245 | 0.38 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr2_-_43605556 | 0.38 |
ENSDART00000084223
|
epc1b
|
enhancer of polycomb homolog 1 (Drosophila) b |
| chr6_+_112579 | 0.38 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr21_-_12749501 | 0.37 |
ENSDART00000179724
|
LO018011.1
|
|
| chr12_-_7280551 | 0.36 |
ENSDART00000061633
|
zgc:171971
|
zgc:171971 |
| chr23_-_5719453 | 0.36 |
ENSDART00000033093
|
lad1
|
ladinin |
| chr17_-_42492668 | 0.36 |
ENSDART00000183946
|
CR925755.2
|
|
| chr12_-_18483348 | 0.35 |
ENSDART00000152757
|
tex2l
|
testis expressed 2, like |
| chr16_+_6864608 | 0.35 |
ENSDART00000078306
|
arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr12_+_13652747 | 0.35 |
ENSDART00000066359
|
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr9_+_54686686 | 0.35 |
ENSDART00000066198
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr24_-_982443 | 0.34 |
ENSDART00000063151
|
napga
|
N-ethylmaleimide-sensitive factor attachment protein, gamma a |
| chr3_-_39627412 | 0.34 |
ENSDART00000123292
|
si:dkey-27o4.1
|
si:dkey-27o4.1 |
| chr20_-_2355357 | 0.34 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr3_-_18805225 | 0.33 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr7_-_8324927 | 0.33 |
ENSDART00000102535
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr17_+_43629008 | 0.33 |
ENSDART00000184185
ENSDART00000181681 |
znf365
|
zinc finger protein 365 |
| chr23_+_35847200 | 0.33 |
ENSDART00000129222
|
rarga
|
retinoic acid receptor gamma a |
| chr22_+_94692 | 0.32 |
ENSDART00000114777
|
nckipsd
|
NCK interacting protein with SH3 domain |
| chr21_+_40498628 | 0.32 |
ENSDART00000163454
|
coro6
|
coronin 6 |
| chr22_-_38360205 | 0.32 |
ENSDART00000162055
|
mark1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr18_-_19103929 | 0.31 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr4_-_38476901 | 0.31 |
ENSDART00000167124
|
si:ch211-209n20.59
|
si:ch211-209n20.59 |
| chr1_-_9134045 | 0.31 |
ENSDART00000142132
|
palb2
|
partner and localizer of BRCA2 |
| chr13_-_280827 | 0.31 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr13_+_7575563 | 0.31 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr7_+_6969909 | 0.31 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr13_-_33114933 | 0.31 |
ENSDART00000140543
ENSDART00000075953 |
zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr13_-_40120252 | 0.30 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr21_-_20765338 | 0.30 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr24_+_42074143 | 0.30 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr22_-_3595439 | 0.30 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr7_-_13381129 | 0.30 |
ENSDART00000164326
|
si:ch73-119p20.1
|
si:ch73-119p20.1 |
| chr12_-_2800809 | 0.30 |
ENSDART00000152682
ENSDART00000083784 |
ubtd1b
|
ubiquitin domain containing 1b |
| chr1_+_55755304 | 0.29 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr15_-_41807371 | 0.29 |
ENSDART00000156819
|
slc25a36b
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36b |
| chr17_-_44440832 | 0.29 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr18_-_1414760 | 0.29 |
ENSDART00000171881
|
PEPD
|
peptidase D |
| chr21_-_14251306 | 0.29 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr7_-_41812015 | 0.29 |
ENSDART00000174058
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr18_+_44703343 | 0.28 |
ENSDART00000131510
|
b3gnt2l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, like |
| chr10_-_35220285 | 0.28 |
ENSDART00000180439
|
ypel2a
|
yippee-like 2a |
| chr8_-_6943155 | 0.28 |
ENSDART00000139545
ENSDART00000033294 |
wdr13
|
WD repeat domain 13 |
| chr22_-_94519 | 0.28 |
ENSDART00000161432
|
ndufaf3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr19_-_38830582 | 0.28 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr22_-_5099824 | 0.28 |
ENSDART00000122341
ENSDART00000161345 |
zfr2
|
zinc finger RNA binding protein 2 |
| chr21_+_11415224 | 0.27 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr22_+_15979430 | 0.27 |
ENSDART00000189703
ENSDART00000192674 |
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr8_-_42712573 | 0.27 |
ENSDART00000132229
ENSDART00000137258 |
si:ch73-138n13.1
|
si:ch73-138n13.1 |
| chr23_-_36418059 | 0.26 |
ENSDART00000135232
|
znf740b
|
zinc finger protein 740b |
| chr9_-_24413008 | 0.25 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr5_+_63749368 | 0.25 |
ENSDART00000135097
|
si:ch73-274k23.1
|
si:ch73-274k23.1 |
| chr12_-_44010532 | 0.25 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr19_-_32944050 | 0.25 |
ENSDART00000137611
|
azin1b
|
antizyme inhibitor 1b |
| chr16_+_10841163 | 0.24 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr5_-_26247671 | 0.24 |
ENSDART00000145187
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr2_-_44971551 | 0.24 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr24_-_42090635 | 0.24 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr3_+_42923275 | 0.24 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr7_-_72426484 | 0.24 |
ENSDART00000190063
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr12_+_34827808 | 0.23 |
ENSDART00000105533
|
tepsin
|
TEPSIN, adaptor related protein complex 4 accessory protein |
| chr18_+_19990412 | 0.23 |
ENSDART00000155054
ENSDART00000090310 |
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr15_-_45356635 | 0.23 |
ENSDART00000192000
|
CABZ01068246.1
|
|
| chr22_+_980290 | 0.23 |
ENSDART00000065377
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr19_-_5103313 | 0.23 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr6_+_45918981 | 0.23 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr10_-_1320114 | 0.22 |
ENSDART00000073617
|
opn4xa
|
opsin 4xa |
| chr9_-_41784799 | 0.22 |
ENSDART00000144573
ENSDART00000112542 ENSDART00000190486 |
obsl1b
|
obscurin-like 1b |
| chr22_-_188102 | 0.22 |
ENSDART00000125391
ENSDART00000170463 |
CABZ01052268.1
|
|
| chr11_-_2478374 | 0.21 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr1_+_19943803 | 0.21 |
ENSDART00000164661
|
apbb2b
|
amyloid beta (A4) precursor protein-binding, family B, member 2b |
| chr5_-_26247215 | 0.21 |
ENSDART00000136806
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr16_-_31622777 | 0.21 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr24_+_33462800 | 0.21 |
ENSDART00000166666
ENSDART00000050826 |
rmc1
|
regulator of MON1-CCZ1 |
| chr4_-_63769637 | 0.20 |
ENSDART00000158757
|
znf1098
|
zinc finger protein 1098 |
| chr14_+_25465346 | 0.20 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr7_-_46019756 | 0.20 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr10_-_16028082 | 0.19 |
ENSDART00000122540
|
aldh7a1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr11_+_42482920 | 0.19 |
ENSDART00000160937
|
arf4a
|
ADP-ribosylation factor 4a |
| chr22_-_29083070 | 0.19 |
ENSDART00000104812
ENSDART00000172576 |
cbx6a
|
chromobox homolog 6a |
| chr24_+_32472155 | 0.19 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr3_-_1859198 | 0.18 |
ENSDART00000121870
|
micall1b.2
|
MICAL like 1b, duplicate 2 |
| chr15_+_8335600 | 0.18 |
ENSDART00000190066
ENSDART00000152603 |
egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr3_-_48993580 | 0.18 |
ENSDART00000182400
|
BX769173.1
|
|
| chr24_-_8777781 | 0.18 |
ENSDART00000082362
ENSDART00000177400 |
mak
|
male germ cell-associated kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of arntl1a+arntl1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.3 | 1.0 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.3 | 1.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.6 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.2 | 0.5 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.2 | 1.6 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.2 | 0.6 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.4 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.5 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.6 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.5 | GO:2000622 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.1 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.1 | 0.8 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.7 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 1.0 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.8 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.0 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 1.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.7 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.6 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.2 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.7 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 1.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.5 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.3 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.8 | GO:0061462 | protein stabilization(GO:0050821) protein localization to lysosome(GO:0061462) |
| 0.0 | 0.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 2.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.7 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.6 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.5 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0030428 | cell septum(GO:0030428) |
| 0.3 | 0.8 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 1.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 1.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.2 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 1.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.0 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.5 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.2 | 0.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 0.7 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 1.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.0 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.1 | 0.3 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.3 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.3 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.5 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 1.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.8 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 1.1 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.3 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 1.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |