Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
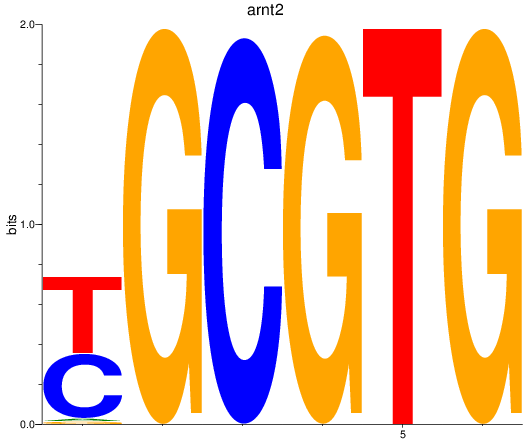
Results for arnt2
Z-value: 2.17
Transcription factors associated with arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arnt2
|
ENSDARG00000103697 | aryl-hydrocarbon receptor nuclear translocator 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arnt2 | dr11_v1_chr7_+_10701938_10701938 | 0.64 | 3.0e-03 | Click! |
Activity profile of arnt2 motif
Sorted Z-values of arnt2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_46313135 | 8.67 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr7_-_74090168 | 8.50 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr14_+_46313396 | 8.35 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr5_-_23429228 | 7.88 |
ENSDART00000049291
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr23_-_6641223 | 7.50 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr5_+_55626693 | 5.54 |
ENSDART00000168908
ENSDART00000161412 |
ntrk2b
|
neurotrophic tyrosine kinase, receptor, type 2b |
| chr17_+_10566490 | 5.53 |
ENSDART00000144408
ENSDART00000137469 |
mgaa
|
MGA, MAX dimerization protein a |
| chr12_-_22540943 | 5.10 |
ENSDART00000172310
|
zbtb4
|
zinc finger and BTB domain containing 4 |
| chr21_+_30194904 | 4.52 |
ENSDART00000137023
ENSDART00000078403 |
BRD8
|
si:ch211-59d17.3 |
| chr11_+_7324704 | 4.38 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr5_+_32162684 | 4.33 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr20_+_35382482 | 4.25 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr5_+_36932718 | 4.23 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr21_+_6751405 | 4.19 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr5_+_23118470 | 4.18 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr13_+_27314795 | 4.17 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr11_+_30513656 | 4.14 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr24_-_11325849 | 4.07 |
ENSDART00000182485
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr10_+_29963518 | 3.82 |
ENSDART00000011317
ENSDART00000099964 ENSDART00000182990 ENSDART00000113912 |
ntm
|
neurotrimin |
| chr20_-_47731768 | 3.73 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr1_+_14283692 | 3.67 |
ENSDART00000017679
|
ppp2r2ca
|
protein phosphatase 2, regulatory subunit B, gamma a |
| chr14_+_41409697 | 3.66 |
ENSDART00000173335
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr2_+_49522178 | 3.59 |
ENSDART00000056254
|
stap2a
|
signal transducing adaptor family member 2a |
| chr17_-_17447899 | 3.59 |
ENSDART00000156928
ENSDART00000109034 |
nrxn3a
|
neurexin 3a |
| chr2_+_42724404 | 3.52 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr5_+_36611128 | 3.46 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr18_-_46208581 | 3.45 |
ENSDART00000141278
|
si:ch211-14c7.2
|
si:ch211-14c7.2 |
| chr15_-_34418525 | 3.36 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr15_+_38859648 | 3.31 |
ENSDART00000141086
|
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr15_-_12319065 | 3.27 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_24428237 | 3.20 |
ENSDART00000189107
ENSDART00000103752 |
dvl1b
|
dishevelled segment polarity protein 1b |
| chr11_+_36989696 | 3.17 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr1_-_22757145 | 3.14 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr12_+_35119762 | 3.13 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr7_-_38340674 | 3.12 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr23_-_8373676 | 3.04 |
ENSDART00000105135
ENSDART00000158531 |
oprl1
|
opiate receptor-like 1 |
| chr21_+_6751760 | 3.04 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr4_+_9279784 | 3.04 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr6_-_39006449 | 3.00 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr17_-_22067451 | 2.91 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr25_+_13406069 | 2.90 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr20_-_8419057 | 2.84 |
ENSDART00000145841
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr1_+_42225060 | 2.84 |
ENSDART00000138740
ENSDART00000101306 |
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr5_-_16983336 | 2.83 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr21_+_6212844 | 2.82 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr23_+_17220986 | 2.73 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr21_-_14630831 | 2.73 |
ENSDART00000132142
ENSDART00000089967 |
cacna1bb
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, b |
| chr13_-_3516473 | 2.72 |
ENSDART00000146240
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr8_-_18899427 | 2.71 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr15_+_36156986 | 2.69 |
ENSDART00000059791
|
sst1.1
|
somatostatin 1, tandem duplicate 1 |
| chr23_-_16692312 | 2.67 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr21_+_28958471 | 2.63 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr21_+_5169154 | 2.59 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr6_+_4872883 | 2.57 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr16_-_13388821 | 2.57 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr1_-_22512063 | 2.57 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr3_+_14768364 | 2.55 |
ENSDART00000090235
ENSDART00000139001 |
nfixb
|
nuclear factor I/Xb |
| chr19_+_17259912 | 2.55 |
ENSDART00000078951
|
MANEAL
|
si:ch211-30b16.2 |
| chr25_+_19954576 | 2.49 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr7_+_10701938 | 2.48 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr1_-_19237576 | 2.45 |
ENSDART00000143663
ENSDART00000131579 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr19_+_49721 | 2.43 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr1_-_8917902 | 2.42 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr19_-_10243148 | 2.40 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr5_-_22952156 | 2.39 |
ENSDART00000111146
|
si:ch211-26b3.4
|
si:ch211-26b3.4 |
| chr18_+_49411417 | 2.33 |
ENSDART00000028944
|
LRFN3
|
zmp:0000001073 |
| chr17_+_9310259 | 2.32 |
ENSDART00000186158
ENSDART00000190329 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr9_-_23217196 | 2.32 |
ENSDART00000083567
|
kif5c
|
kinesin family member 5C |
| chr19_+_15571290 | 2.32 |
ENSDART00000131134
|
foxo6b
|
forkhead box O6 b |
| chr19_-_1023051 | 2.27 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr5_-_32092856 | 2.26 |
ENSDART00000086181
ENSDART00000181677 |
cabp7b
|
calcium binding protein 7b |
| chr13_+_24022963 | 2.25 |
ENSDART00000028285
|
pgbd5
|
piggyBac transposable element derived 5 |
| chr1_+_36436936 | 2.24 |
ENSDART00000124112
|
pou4f2
|
POU class 4 homeobox 2 |
| chr14_+_8275115 | 2.24 |
ENSDART00000129055
|
nrg2b
|
neuregulin 2b |
| chr11_+_41135055 | 2.21 |
ENSDART00000173252
|
camta1
|
calmodulin binding transcription activator 1 |
| chr6_-_26559921 | 2.21 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr17_-_20287530 | 2.19 |
ENSDART00000078703
ENSDART00000191289 |
add3b
|
adducin 3 (gamma) b |
| chr19_+_48176745 | 2.19 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr21_+_6780340 | 2.18 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr14_-_47314340 | 2.17 |
ENSDART00000164851
|
fstl5
|
follistatin-like 5 |
| chr6_-_14292307 | 2.16 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr2_-_24603325 | 2.16 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr1_+_33969015 | 2.16 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr16_+_10918252 | 2.15 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr18_-_33344 | 2.15 |
ENSDART00000129125
|
pde8a
|
phosphodiesterase 8A |
| chr11_-_36963988 | 2.14 |
ENSDART00000168288
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr1_-_22756898 | 2.11 |
ENSDART00000158915
|
prom1b
|
prominin 1 b |
| chr8_+_1082100 | 2.05 |
ENSDART00000149276
|
lzts3b
|
leucine zipper, putative tumor suppressor family member 3b |
| chr13_+_12299997 | 2.00 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr3_+_18097700 | 1.98 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr1_-_18446161 | 1.98 |
ENSDART00000089442
|
klhl5
|
kelch-like family member 5 |
| chr7_-_44963154 | 1.96 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr9_-_44295071 | 1.95 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr15_-_12229874 | 1.95 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr22_-_14128716 | 1.95 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr11_-_37589293 | 1.93 |
ENSDART00000172989
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr4_-_4780667 | 1.92 |
ENSDART00000133973
|
si:ch211-258f14.2
|
si:ch211-258f14.2 |
| chr23_-_12158685 | 1.90 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr19_-_205104 | 1.90 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr8_+_10561922 | 1.89 |
ENSDART00000133348
|
fam19a5l
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5-like |
| chr19_+_10396042 | 1.87 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr11_-_3959477 | 1.87 |
ENSDART00000045971
|
pbrm1
|
polybromo 1 |
| chr12_-_431249 | 1.83 |
ENSDART00000083827
|
hs3st3l
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3-like |
| chr20_+_826459 | 1.81 |
ENSDART00000104740
|
nt5e
|
5'-nucleotidase, ecto (CD73) |
| chr12_+_9703172 | 1.79 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr14_-_7885707 | 1.78 |
ENSDART00000029981
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr11_+_30161699 | 1.78 |
ENSDART00000190504
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr13_-_30027730 | 1.78 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr7_+_25858380 | 1.76 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr2_+_21090317 | 1.75 |
ENSDART00000109568
ENSDART00000139633 |
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr11_+_34824099 | 1.70 |
ENSDART00000037017
ENSDART00000146944 |
slc38a3a
|
solute carrier family 38, member 3a |
| chr2_+_47581997 | 1.70 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr1_-_7917062 | 1.69 |
ENSDART00000177068
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr16_-_33059246 | 1.67 |
ENSDART00000171718
ENSDART00000168305 ENSDART00000166401 |
snap91
|
synaptosomal-associated protein 91 |
| chr11_+_30161168 | 1.66 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr8_-_23328264 | 1.66 |
ENSDART00000131934
|
nol4la
|
nucleolar protein 4-like a |
| chr15_-_28247583 | 1.66 |
ENSDART00000112967
|
rilp
|
Rab interacting lysosomal protein |
| chr17_+_28340138 | 1.65 |
ENSDART00000033943
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr5_-_71460556 | 1.64 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr22_-_29191152 | 1.64 |
ENSDART00000132702
|
pvalb7
|
parvalbumin 7 |
| chr16_-_13662514 | 1.64 |
ENSDART00000146348
|
shisa7a
|
shisa family member 7a |
| chr4_-_27350820 | 1.63 |
ENSDART00000145806
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr3_+_17744339 | 1.62 |
ENSDART00000132622
|
znf385c
|
zinc finger protein 385C |
| chr6_+_11829867 | 1.62 |
ENSDART00000151044
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr4_-_4706893 | 1.61 |
ENSDART00000093005
|
CABZ01020835.1
|
|
| chr24_-_38110779 | 1.61 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr24_-_18179535 | 1.60 |
ENSDART00000186112
|
cntnap2a
|
contactin associated protein like 2a |
| chr24_-_3419998 | 1.59 |
ENSDART00000066839
|
slc35g2b
|
solute carrier family 35, member G2b |
| chr19_+_33732188 | 1.59 |
ENSDART00000151192
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr10_-_2682198 | 1.58 |
ENSDART00000183727
|
pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr25_+_4541211 | 1.55 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr19_+_42660158 | 1.55 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr5_+_3501859 | 1.54 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr3_-_3496738 | 1.54 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr6_-_58910402 | 1.53 |
ENSDART00000156662
|
mbd6
|
methyl-CpG binding domain protein 6 |
| chr8_-_11640240 | 1.53 |
ENSDART00000091752
|
fnbp1a
|
formin binding protein 1a |
| chr22_-_21150845 | 1.53 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr16_+_1802307 | 1.52 |
ENSDART00000180026
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr1_+_8662530 | 1.52 |
ENSDART00000054989
|
fscn1b
|
fascin actin-bundling protein 1b |
| chr1_+_31942961 | 1.50 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr2_+_25929619 | 1.50 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr1_+_42224769 | 1.50 |
ENSDART00000177496
ENSDART00000184778 ENSDART00000110860 |
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr4_+_7888047 | 1.49 |
ENSDART00000104676
|
camk1da
|
calcium/calmodulin-dependent protein kinase 1Da |
| chr4_-_2867461 | 1.49 |
ENSDART00000160308
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr21_-_27185915 | 1.49 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr19_-_10395683 | 1.48 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr15_-_33734105 | 1.48 |
ENSDART00000172729
ENSDART00000172045 |
trmt9b
|
tRNA methyltransferase 9B |
| chr10_-_15053507 | 1.48 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr1_-_45633955 | 1.47 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr6_+_40992409 | 1.45 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr5_-_28915130 | 1.44 |
ENSDART00000078592
|
npdc1b
|
neural proliferation, differentiation and control, 1b |
| chr16_-_12953739 | 1.44 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr13_-_17464362 | 1.43 |
ENSDART00000145499
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr4_+_10888762 | 1.43 |
ENSDART00000136049
|
syt10
|
synaptotagmin X |
| chr19_-_47452874 | 1.42 |
ENSDART00000025931
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr8_+_20624510 | 1.42 |
ENSDART00000138604
|
nfic
|
nuclear factor I/C |
| chr21_-_41305748 | 1.41 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr8_+_44623540 | 1.40 |
ENSDART00000141513
|
grk5l
|
G protein-coupled receptor kinase 5 like |
| chr23_-_44494401 | 1.40 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr11_+_34824262 | 1.38 |
ENSDART00000103157
|
slc38a3a
|
solute carrier family 38, member 3a |
| chr12_-_25150239 | 1.38 |
ENSDART00000038415
ENSDART00000135368 |
rhoq
|
ras homolog family member Q |
| chr13_-_32648382 | 1.37 |
ENSDART00000138571
ENSDART00000132820 |
bend3
|
BEN domain containing 3 |
| chr3_-_62380146 | 1.36 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr13_-_31166544 | 1.36 |
ENSDART00000146250
ENSDART00000132129 ENSDART00000139591 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr10_+_36650222 | 1.34 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr13_+_12739283 | 1.34 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr24_+_35881984 | 1.34 |
ENSDART00000066587
|
klhl14
|
kelch-like family member 14 |
| chr12_-_14922955 | 1.33 |
ENSDART00000002078
|
neurod2
|
neurogenic differentiation 2 |
| chr25_-_36282539 | 1.32 |
ENSDART00000073398
|
slc7a10b
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10b |
| chr3_+_46628885 | 1.32 |
ENSDART00000006602
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr4_+_121709 | 1.32 |
ENSDART00000186461
ENSDART00000172255 |
crebl2
|
cAMP responsive element binding protein-like 2 |
| chr7_-_33949246 | 1.32 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr23_-_31555696 | 1.31 |
ENSDART00000053539
|
tcf21
|
transcription factor 21 |
| chr17_-_45552602 | 1.31 |
ENSDART00000154844
ENSDART00000034432 |
susd4
|
sushi domain containing 4 |
| chr4_-_4780510 | 1.30 |
ENSDART00000109609
|
si:ch211-258f14.2
|
si:ch211-258f14.2 |
| chr23_+_22656477 | 1.30 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr5_-_48285756 | 1.29 |
ENSDART00000183495
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr25_-_32311048 | 1.29 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr13_-_17464654 | 1.28 |
ENSDART00000140312
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr17_+_22530820 | 1.28 |
ENSDART00000089916
|
edaradd
|
EDAR-associated death domain |
| chr18_-_44610992 | 1.28 |
ENSDART00000125968
ENSDART00000185836 |
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr21_-_435466 | 1.28 |
ENSDART00000110297
|
klf4
|
Kruppel-like factor 4 |
| chr22_-_11054244 | 1.28 |
ENSDART00000105823
|
insrb
|
insulin receptor b |
| chr15_-_43978141 | 1.27 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr20_-_2949028 | 1.26 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr23_+_35847200 | 1.26 |
ENSDART00000129222
|
rarga
|
retinoic acid receptor gamma a |
| chr13_-_20518632 | 1.24 |
ENSDART00000165310
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr14_-_34276574 | 1.23 |
ENSDART00000021437
|
gria1a
|
glutamate receptor, ionotropic, AMPA 1a |
| chr7_-_56793739 | 1.22 |
ENSDART00000082842
|
si:ch211-146m13.3
|
si:ch211-146m13.3 |
| chr21_-_10446405 | 1.22 |
ENSDART00000167948
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr17_+_52823015 | 1.21 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr21_+_23953181 | 1.21 |
ENSDART00000145541
ENSDART00000065599 ENSDART00000112869 |
cadm1a
|
cell adhesion molecule 1a |
| chr16_-_44127307 | 1.20 |
ENSDART00000104583
ENSDART00000151936 ENSDART00000058685 ENSDART00000190830 |
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr4_-_6567355 | 1.20 |
ENSDART00000134820
ENSDART00000142087 |
foxp2
|
forkhead box P2 |
| chr19_+_33732487 | 1.20 |
ENSDART00000010294
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr15_+_28482862 | 1.19 |
ENSDART00000015286
ENSDART00000154320 |
ankrd13b
|
ankyrin repeat domain 13B |
| chr10_-_15919839 | 1.18 |
ENSDART00000065032
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr11_+_6152643 | 1.18 |
ENSDART00000012789
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr19_-_42651615 | 1.18 |
ENSDART00000123360
|
susd5
|
sushi domain containing 5 |
| chr8_-_21184759 | 1.18 |
ENSDART00000139257
|
gls2a
|
glutaminase 2a (liver, mitochondrial) |
Network of associatons between targets according to the STRING database.
First level regulatory network of arnt2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 1.7 | 5.1 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 1.5 | 4.4 | GO:0042940 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 1.4 | 4.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 1.1 | 4.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 1.0 | 4.2 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.9 | 8.5 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.9 | 4.4 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.8 | 7.5 | GO:0006833 | water transport(GO:0006833) |
| 0.8 | 3.1 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.7 | 3.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.6 | 3.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.6 | 1.7 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.5 | 1.5 | GO:0042044 | fluid transport(GO:0042044) |
| 0.5 | 1.4 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.5 | 3.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.5 | 3.2 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.4 | 4.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.4 | 1.8 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 | 1.3 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.4 | 1.3 | GO:2000257 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 0.4 | 1.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 1.6 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.4 | 0.4 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.4 | 2.0 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 1.5 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.4 | 3.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.4 | 5.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.4 | 1.8 | GO:0051701 | interaction with host(GO:0051701) |
| 0.4 | 2.8 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.4 | 1.8 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.3 | 1.4 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.3 | 1.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 2.3 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.3 | 3.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.3 | 1.6 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.3 | 1.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 1.8 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.3 | 5.7 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.3 | 0.9 | GO:0071314 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.3 | 1.2 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.3 | 1.1 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.3 | 1.0 | GO:0015677 | copper ion import(GO:0015677) |
| 0.3 | 0.5 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.3 | 2.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 2.5 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.2 | 1.0 | GO:0021755 | eurydendroid cell differentiation(GO:0021755) |
| 0.2 | 3.0 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.2 | 2.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.2 | 1.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.2 | 2.6 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 1.3 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 2.1 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.2 | 0.6 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.2 | 6.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 1.9 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 0.8 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.2 | 0.6 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 0.5 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.2 | 5.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 2.8 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.2 | 1.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 3.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 1.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 1.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 1.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.2 | 1.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 1.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 0.8 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.2 | 1.4 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.2 | 0.8 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 3.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 2.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 2.4 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.1 | 17.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.6 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.8 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.8 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 0.9 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.2 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 1.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.4 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 4.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.6 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 1.5 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.8 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 1.0 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.6 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 2.8 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.4 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.1 | 0.9 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 4.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 3.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.4 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 0.9 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.0 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 0.6 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 3.0 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 4.0 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.3 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 1.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 1.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.6 | GO:0042461 | photoreceptor cell development(GO:0042461) |
| 0.1 | 1.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.6 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 4.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 1.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 3.3 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.1 | 1.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.7 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 0.6 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 1.3 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 2.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.5 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 3.0 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 1.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 1.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.7 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 0.5 | GO:0010890 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.1 | 0.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.4 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.6 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 2.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 1.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 1.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 2.2 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.6 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.8 | GO:0006775 | fat-soluble vitamin metabolic process(GO:0006775) |
| 0.0 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 1.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 2.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 1.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.0 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 2.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.2 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 1.0 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 1.3 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 1.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.0 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 5.5 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 2.3 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.5 | GO:0038084 | vascular endothelial growth factor signaling pathway(GO:0038084) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 2.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.0 | 0.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.5 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 1.4 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 2.2 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 0.9 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 5.2 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.8 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.3 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 1.3 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.4 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.7 | 4.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.7 | 8.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.7 | 5.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.5 | 1.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.4 | 5.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 16.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 3.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 1.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 2.8 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.2 | 2.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 1.9 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 5.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 1.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 1.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 1.9 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 1.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 3.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 4.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 2.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 5.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.7 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 2.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.5 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 1.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 3.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 17.3 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 3.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 2.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 2.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 3.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 3.5 | GO:0070382 | exocytic vesicle(GO:0070382) secretory vesicle(GO:0099503) |
| 0.0 | 0.4 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.5 | GO:0005770 | late endosome(GO:0005770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.8 | 7.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.7 | 4.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.6 | 9.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.6 | 3.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.5 | 3.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.5 | 3.1 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.5 | 3.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 1.8 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.4 | 2.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.4 | 3.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.4 | 3.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.3 | 1.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 1.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 6.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 2.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 0.9 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.3 | 1.0 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 1.3 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 2.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 17.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.6 | GO:0005330 | dopamine transmembrane transporter activity(GO:0005329) dopamine:sodium symporter activity(GO:0005330) norepinephrine transmembrane transporter activity(GO:0005333) norepinephrine:sodium symporter activity(GO:0005334) |
| 0.2 | 1.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 5.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 0.8 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.2 | 0.6 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.2 | 4.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 1.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 0.9 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 4.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 1.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 1.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.8 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.2 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 3.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 1.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 1.0 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 7.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.7 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 4.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.0 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 9.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.9 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 2.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 1.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 2.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.8 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 2.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.6 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 3.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 2.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.0 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 3.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.2 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 1.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 2.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 2.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 3.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.3 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.1 | 0.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 2.0 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 2.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.0 | 3.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 1.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.0 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 3.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.3 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 1.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.3 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.0 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 1.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 2.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 2.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.4 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 2.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 2.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 2.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 2.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 1.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.3 | 4.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 1.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 1.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 1.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 2.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 1.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |