Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
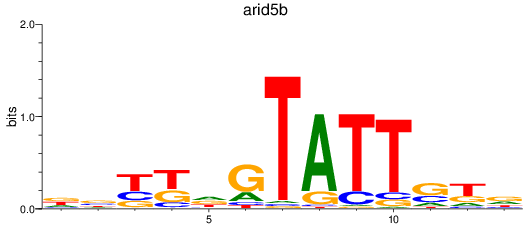
Results for arid5b
Z-value: 0.40
Transcription factors associated with arid5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid5b
|
ENSDARG00000037196 | AT-rich interaction domain 5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid5b | dr11_v1_chr12_+_8168272_8168272 | 0.51 | 2.5e-02 | Click! |
Activity profile of arid5b motif
Sorted Z-values of arid5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_21832441 | 1.03 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr3_+_19299309 | 0.92 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr22_+_19311411 | 0.91 |
ENSDART00000133234
ENSDART00000138284 |
si:dkey-21e2.16
|
si:dkey-21e2.16 |
| chr18_+_5543677 | 0.89 |
ENSDART00000146161
ENSDART00000136189 |
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr9_+_5862040 | 0.85 |
ENSDART00000129117
|
pdzk1
|
PDZ domain containing 1 |
| chr13_-_31166544 | 0.81 |
ENSDART00000146250
ENSDART00000132129 ENSDART00000139591 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr9_-_52814204 | 0.80 |
ENSDART00000140771
ENSDART00000007401 |
MAP3K13
|
si:ch211-45c16.2 |
| chr14_-_46238186 | 0.73 |
ENSDART00000173245
|
si:ch211-113d11.6
|
si:ch211-113d11.6 |
| chr11_+_13224281 | 0.69 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr2_-_58142854 | 0.61 |
ENSDART00000169909
|
CABZ01110881.1
|
|
| chr5_-_69312533 | 0.58 |
ENSDART00000082614
ENSDART00000183098 |
smtnb
|
smoothelin b |
| chr23_+_44665230 | 0.56 |
ENSDART00000144958
|
camta2
|
calmodulin binding transcription activator 2 |
| chr7_+_73780936 | 0.55 |
ENSDART00000092691
|
pea15
|
proliferation and apoptosis adaptor protein 15 |
| chr20_+_54037138 | 0.53 |
ENSDART00000143172
|
wdr20b
|
WD repeat domain 20b |
| chr5_-_38155005 | 0.51 |
ENSDART00000097770
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr10_+_2876020 | 0.47 |
ENSDART00000136618
ENSDART00000133128 ENSDART00000140803 ENSDART00000141505 |
ccar2
|
cell cycle and apoptosis regulator 2 |
| chr25_+_33063762 | 0.43 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr18_+_36782930 | 0.43 |
ENSDART00000004129
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr1_+_9994811 | 0.42 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr2_+_45548890 | 0.42 |
ENSDART00000113994
|
fndc7a
|
fibronectin type III domain containing 7a |
| chr20_+_2460864 | 0.41 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr23_-_29505645 | 0.40 |
ENSDART00000146458
|
kif1b
|
kinesin family member 1B |
| chr12_+_19976400 | 0.39 |
ENSDART00000153177
|
mkl2a
|
MKL/myocardin-like 2a |
| chr7_-_52709759 | 0.38 |
ENSDART00000136745
ENSDART00000131282 ENSDART00000073766 ENSDART00000161387 ENSDART00000174117 ENSDART00000174355 |
tcf12
|
transcription factor 12 |
| chr4_-_20043484 | 0.37 |
ENSDART00000167780
|
zgc:193726
|
zgc:193726 |
| chr15_+_42626957 | 0.36 |
ENSDART00000154754
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr20_+_1278184 | 0.34 |
ENSDART00000039616
|
lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr17_-_50022827 | 0.34 |
ENSDART00000161008
|
filip1a
|
filamin A interacting protein 1a |
| chr16_+_14033121 | 0.33 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr16_+_1254390 | 0.32 |
ENSDART00000092627
|
ADAMTSL4
|
ADAMTS like 4 |
| chr7_+_38510197 | 0.32 |
ENSDART00000173468
ENSDART00000100479 |
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr3_-_36932515 | 0.32 |
ENSDART00000111981
ENSDART00000188470 |
cntnap1
|
contactin associated protein 1 |
| chr24_+_30392834 | 0.30 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr21_-_1625976 | 0.29 |
ENSDART00000066621
|
vmo1b
|
vitelline membrane outer layer 1 homolog b |
| chr3_-_4303262 | 0.29 |
ENSDART00000112819
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr10_-_29101715 | 0.29 |
ENSDART00000149674
ENSDART00000171194 ENSDART00000192019 |
si:ch211-103f14.3
|
si:ch211-103f14.3 |
| chr23_-_24047054 | 0.28 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr19_-_45650994 | 0.27 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr16_+_41570653 | 0.27 |
ENSDART00000102665
|
aste1a
|
asteroid homolog 1a |
| chr17_-_51224800 | 0.27 |
ENSDART00000150089
|
psen1
|
presenilin 1 |
| chr2_+_42871831 | 0.24 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr22_-_19102256 | 0.24 |
ENSDART00000171866
ENSDART00000166295 |
polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr6_+_13933464 | 0.22 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr23_-_27101600 | 0.22 |
ENSDART00000139231
|
stat6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr4_+_61840440 | 0.22 |
ENSDART00000169427
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr2_+_42072231 | 0.22 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr12_-_25097520 | 0.22 |
ENSDART00000158036
|
cript
|
cysteine-rich PDZ-binding protein |
| chr18_+_3052408 | 0.21 |
ENSDART00000181563
|
kctd14
|
potassium channel tetramerization domain containing 14 |
| chr9_-_14992730 | 0.21 |
ENSDART00000137117
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr4_+_54618332 | 0.20 |
ENSDART00000171824
|
si:ch211-227e10.2
|
si:ch211-227e10.2 |
| chr12_+_13652361 | 0.20 |
ENSDART00000182757
ENSDART00000152689 |
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr1_+_7956030 | 0.20 |
ENSDART00000159655
|
CR855320.1
|
|
| chr24_-_6644734 | 0.20 |
ENSDART00000167391
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr5_+_64277604 | 0.20 |
ENSDART00000111282
|
qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr23_+_45734011 | 0.20 |
ENSDART00000062415
|
CABZ01088025.1
|
|
| chr12_-_46252062 | 0.19 |
ENSDART00000153223
|
si:ch211-226h7.5
|
si:ch211-226h7.5 |
| chr24_-_6546479 | 0.19 |
ENSDART00000160538
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr19_-_32042105 | 0.19 |
ENSDART00000088358
|
znf704
|
zinc finger protein 704 |
| chr5_-_29112956 | 0.18 |
ENSDART00000132726
|
whrnb
|
whirlin b |
| chr10_-_35410518 | 0.18 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr13_-_4704353 | 0.17 |
ENSDART00000171900
|
oit3
|
oncoprotein induced transcript 3 |
| chr8_+_36500061 | 0.17 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr3_-_10634438 | 0.17 |
ENSDART00000093037
ENSDART00000130761 ENSDART00000156617 |
map2k4a
|
mitogen-activated protein kinase kinase 4a |
| chr24_+_31277360 | 0.17 |
ENSDART00000165993
|
f3a
|
coagulation factor IIIa |
| chr22_+_21972824 | 0.17 |
ENSDART00000183708
ENSDART00000133739 |
gna15.3
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 3 |
| chr21_+_39365920 | 0.16 |
ENSDART00000140940
|
cluhb
|
clustered mitochondria (cluA/CLU1) homolog b |
| chr7_+_57109214 | 0.16 |
ENSDART00000135068
ENSDART00000098412 |
enosf1
|
enolase superfamily member 1 |
| chr13_+_40034176 | 0.16 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr18_+_31410652 | 0.15 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr8_+_8973425 | 0.15 |
ENSDART00000066107
|
bcap31
|
B cell receptor associated protein 31 |
| chr1_+_37195465 | 0.15 |
ENSDART00000043855
ENSDART00000192580 ENSDART00000181666 |
dclk2a
|
doublecortin-like kinase 2a |
| chr17_+_10578823 | 0.15 |
ENSDART00000134610
|
mgaa
|
MGA, MAX dimerization protein a |
| chr14_+_8940326 | 0.14 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr16_-_25606235 | 0.14 |
ENSDART00000192741
|
zgc:110410
|
zgc:110410 |
| chr12_+_20618336 | 0.14 |
ENSDART00000084007
|
si:dkey-220f10.4
|
si:dkey-220f10.4 |
| chr1_+_37195919 | 0.13 |
ENSDART00000159684
ENSDART00000172742 ENSDART00000158395 |
dclk2a
|
doublecortin-like kinase 2a |
| chr3_+_28953274 | 0.12 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr23_-_15878879 | 0.12 |
ENSDART00000010119
|
eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr4_-_815871 | 0.12 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr10_-_28193642 | 0.11 |
ENSDART00000019050
|
rps6kb1a
|
ribosomal protein S6 kinase b, polypeptide 1a |
| chr8_-_18532957 | 0.11 |
ENSDART00000192783
ENSDART00000190530 |
nexn
|
nexilin (F actin binding protein) |
| chr21_+_3928947 | 0.11 |
ENSDART00000149777
|
setx
|
senataxin |
| chr8_+_36500308 | 0.11 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr2_+_42072689 | 0.10 |
ENSDART00000134203
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr22_-_10586191 | 0.10 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr24_-_6078222 | 0.10 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr12_+_20618522 | 0.10 |
ENSDART00000091452
|
si:dkey-220f10.4
|
si:dkey-220f10.4 |
| chr20_+_12702923 | 0.10 |
ENSDART00000163499
|
zgc:153383
|
zgc:153383 |
| chr12_-_46315898 | 0.10 |
ENSDART00000153138
|
si:ch211-226h7.3
|
si:ch211-226h7.3 |
| chr6_+_58406014 | 0.10 |
ENSDART00000044241
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr21_-_2322102 | 0.09 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr7_+_17716601 | 0.09 |
ENSDART00000173792
ENSDART00000080825 |
rtn3
|
reticulon 3 |
| chr16_+_13818500 | 0.09 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr15_-_47812268 | 0.08 |
ENSDART00000190231
|
tomt
|
transmembrane O-methyltransferase |
| chr9_+_14023386 | 0.08 |
ENSDART00000140199
ENSDART00000124267 |
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr9_+_29985010 | 0.08 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr11_-_43002262 | 0.08 |
ENSDART00000172477
ENSDART00000181513 |
CABZ01069998.1
|
|
| chr8_+_47342586 | 0.08 |
ENSDART00000007624
|
plch2a
|
phospholipase C, eta 2a |
| chr13_+_22421151 | 0.07 |
ENSDART00000122687
|
opn4a
|
opsin 4a (melanopsin) |
| chr14_-_21238046 | 0.07 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr13_+_30215765 | 0.07 |
ENSDART00000041026
ENSDART00000133049 |
qrfpra
|
pyroglutamylated RFamide peptide receptor a |
| chr25_+_35133404 | 0.07 |
ENSDART00000188505
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr10_-_18463934 | 0.06 |
ENSDART00000133116
ENSDART00000113422 |
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr9_+_21281652 | 0.06 |
ENSDART00000113352
|
lats2
|
large tumor suppressor kinase 2 |
| chr19_-_24218942 | 0.06 |
ENSDART00000189198
|
BX547993.2
|
|
| chr3_+_23047241 | 0.06 |
ENSDART00000103858
|
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr13_+_35635672 | 0.06 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr1_+_58066109 | 0.05 |
ENSDART00000115190
|
si:ch211-114l13.4
|
si:ch211-114l13.4 |
| chr18_+_27439680 | 0.05 |
ENSDART00000014726
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr12_+_46841918 | 0.05 |
ENSDART00000157464
|
adkb
|
adenosine kinase b |
| chr4_+_53119675 | 0.05 |
ENSDART00000167470
|
si:dkey-8o9.2
|
si:dkey-8o9.2 |
| chr4_-_32458327 | 0.05 |
ENSDART00000182527
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr11_+_42765963 | 0.05 |
ENSDART00000156080
ENSDART00000179888 |
tdrd3
|
tudor domain containing 3 |
| chr20_-_49657134 | 0.04 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr13_+_8987957 | 0.04 |
ENSDART00000148144
|
hcar1-3
|
hydroxycarboxylic acid receptor 1-3 |
| chr23_-_24542156 | 0.04 |
ENSDART00000132265
|
atp13a2
|
ATPase 13A2 |
| chr4_-_76205022 | 0.04 |
ENSDART00000159687
ENSDART00000187593 |
si:ch211-106j21.5
|
si:ch211-106j21.5 |
| chr1_+_58602506 | 0.03 |
ENSDART00000158425
|
si:ch73-221f6.1
|
si:ch73-221f6.1 |
| chr19_-_11977783 | 0.03 |
ENSDART00000183337
|
si:ch1073-296d18.1
|
si:ch1073-296d18.1 |
| chr21_-_38582061 | 0.03 |
ENSDART00000193891
ENSDART00000124349 |
pof1b
|
POF1B, actin binding protein |
| chr10_+_8481660 | 0.03 |
ENSDART00000109559
|
BX682234.1
|
|
| chr16_-_42303856 | 0.02 |
ENSDART00000180030
|
ppox
|
protoporphyrinogen oxidase |
| chr23_+_6926183 | 0.02 |
ENSDART00000149882
|
si:ch211-117c9.1
|
si:ch211-117c9.1 |
| chr25_+_9027831 | 0.02 |
ENSDART00000155855
|
im:7145024
|
im:7145024 |
| chr4_-_71151245 | 0.02 |
ENSDART00000165415
|
si:dkey-193i10.4
|
si:dkey-193i10.4 |
| chr20_+_51466046 | 0.02 |
ENSDART00000178065
|
tlr5b
|
toll-like receptor 5b |
| chr15_+_23356600 | 0.01 |
ENSDART00000113279
|
rnf26
|
ring finger protein 26 |
| chr14_+_20351 | 0.01 |
ENSDART00000051893
|
stx18
|
syntaxin 18 |
| chr4_+_49196098 | 0.01 |
ENSDART00000150678
|
si:dkey-40n15.1
|
si:dkey-40n15.1 |
| chr1_-_633356 | 0.01 |
ENSDART00000171019
|
appa
|
amyloid beta (A4) precursor protein a |
| chr4_+_20177526 | 0.01 |
ENSDART00000017947
ENSDART00000135451 |
ccdc146
|
coiled-coil domain containing 146 |
| chr11_-_33960318 | 0.01 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr16_+_53387085 | 0.01 |
ENSDART00000154223
ENSDART00000101404 |
kif13a
|
kinesin family member 13A |
| chr9_+_29603649 | 0.00 |
ENSDART00000140477
|
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr11_+_42726712 | 0.00 |
ENSDART00000028955
|
tdrd3
|
tudor domain containing 3 |
| chr22_+_19478140 | 0.00 |
ENSDART00000135291
ENSDART00000136576 |
si:dkey-78l4.8
|
si:dkey-78l4.8 |
| chr25_+_32157326 | 0.00 |
ENSDART00000112588
|
tjp1b
|
tight junction protein 1b |
| chr5_-_69523816 | 0.00 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of arid5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.2 | 0.7 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.2 | 0.9 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.4 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.1 | 0.3 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.3 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 0.3 | GO:0019860 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.1 | 0.3 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.3 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 1.0 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.6 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.1 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 1.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.2 | 0.9 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.2 | 0.7 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |