Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
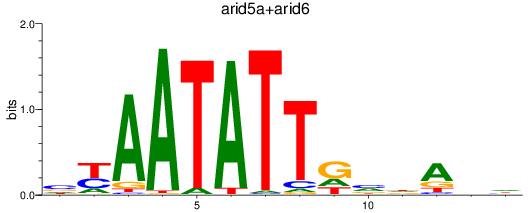
Results for arid5a+arid6
Z-value: 0.64
Transcription factors associated with arid5a+arid6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid6
|
ENSDARG00000069988 | AT-rich interaction domain 6 |
|
arid5a
|
ENSDARG00000077120 | AT rich interactive domain 5A (MRF1-like) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid6 | dr11_v1_chr21_+_11244068_11244068 | 0.12 | 6.2e-01 | Click! |
| arid5a | dr11_v1_chr8_+_52377516_52377516 | 0.07 | 7.7e-01 | Click! |
Activity profile of arid5a+arid6 motif
Sorted Z-values of arid5a+arid6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_53344726 | 0.89 |
ENSDART00000184395
ENSDART00000170212 |
CU914536.1
|
|
| chr4_+_15944245 | 0.77 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr23_+_5631381 | 0.77 |
ENSDART00000149143
|
pkp1a
|
plakophilin 1a |
| chr2_-_44280061 | 0.73 |
ENSDART00000136818
|
mpz
|
myelin protein zero |
| chr4_-_8043839 | 0.72 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr10_+_15967341 | 0.67 |
ENSDART00000115130
ENSDART00000189687 |
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr19_-_1948236 | 0.67 |
ENSDART00000163344
|
znrf2a
|
zinc and ring finger 2a |
| chr3_-_38915998 | 0.65 |
ENSDART00000141886
|
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr20_-_9755546 | 0.61 |
ENSDART00000152498
ENSDART00000152239 |
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr23_+_383782 | 0.60 |
ENSDART00000055148
|
zgc:101663
|
zgc:101663 |
| chr21_+_36623162 | 0.54 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr6_+_45932276 | 0.54 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr24_-_11446156 | 0.54 |
ENSDART00000143921
ENSDART00000066778 |
acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr21_-_2261720 | 0.52 |
ENSDART00000170161
|
si:ch73-299h12.2
|
si:ch73-299h12.2 |
| chr22_+_15343953 | 0.51 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr22_-_606067 | 0.50 |
ENSDART00000136722
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr21_+_20903244 | 0.50 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr6_-_8466717 | 0.49 |
ENSDART00000151577
ENSDART00000151800 ENSDART00000151227 |
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr10_+_38526496 | 0.49 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr10_+_44699734 | 0.48 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr2_+_21048661 | 0.47 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr4_+_12031958 | 0.47 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr5_+_13427826 | 0.46 |
ENSDART00000083359
|
sec14l8
|
SEC14-like lipid binding 8 |
| chr19_+_21820144 | 0.46 |
ENSDART00000181996
|
CU856623.1
|
|
| chr17_-_24564674 | 0.45 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr3_+_3641429 | 0.43 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr1_+_29068654 | 0.43 |
ENSDART00000053932
|
cbsa
|
cystathionine-beta-synthase a |
| chr13_-_12021566 | 0.42 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr16_+_26777473 | 0.42 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr19_-_1947403 | 0.41 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr7_+_20467549 | 0.40 |
ENSDART00000173724
|
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr14_-_34700633 | 0.40 |
ENSDART00000150358
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr20_+_1960092 | 0.39 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr1_-_50838160 | 0.38 |
ENSDART00000163939
ENSDART00000165111 |
zgc:154142
|
zgc:154142 |
| chr12_+_2665081 | 0.38 |
ENSDART00000147532
|
rbp3
|
retinol binding protein 3 |
| chr7_+_36484598 | 0.37 |
ENSDART00000173595
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr5_-_66429357 | 0.36 |
ENSDART00000160189
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr7_-_30925798 | 0.36 |
ENSDART00000149303
|
sord
|
sorbitol dehydrogenase |
| chr2_-_20715094 | 0.36 |
ENSDART00000155439
|
dusp12
|
dual specificity phosphatase 12 |
| chr10_-_43655449 | 0.36 |
ENSDART00000099134
|
mef2ca
|
myocyte enhancer factor 2ca |
| chr10_+_45089820 | 0.36 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr24_-_36238054 | 0.36 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr8_+_29636431 | 0.36 |
ENSDART00000133047
|
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr18_+_15876385 | 0.36 |
ENSDART00000142527
|
eea1
|
early endosome antigen 1 |
| chr16_-_4836218 | 0.35 |
ENSDART00000054077
|
tpbgb
|
trophoblast glycoprotein b |
| chr20_+_46925581 | 0.35 |
ENSDART00000192531
|
si:ch73-21k16.5
|
si:ch73-21k16.5 |
| chr1_+_32054159 | 0.34 |
ENSDART00000181442
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr4_+_42604252 | 0.34 |
ENSDART00000184850
|
CR925768.1
|
|
| chr1_+_21731382 | 0.34 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr13_-_28689717 | 0.34 |
ENSDART00000101582
|
pcgf6
|
polycomb group ring finger 6 |
| chr9_+_52492639 | 0.34 |
ENSDART00000078939
|
march4
|
membrane-associated ring finger (C3HC4) 4 |
| chr11_+_25504215 | 0.34 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr5_-_42059869 | 0.34 |
ENSDART00000193984
|
cenpv
|
centromere protein V |
| chr23_+_45512825 | 0.33 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
| chr22_+_36656680 | 0.33 |
ENSDART00000134031
ENSDART00000056169 |
b3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr19_-_48336535 | 0.33 |
ENSDART00000162752
|
si:ch73-359m17.6
|
si:ch73-359m17.6 |
| chr2_-_57110477 | 0.33 |
ENSDART00000181132
|
slc25a42
|
solute carrier family 25, member 42 |
| chr9_+_37329036 | 0.33 |
ENSDART00000131756
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr7_+_39399747 | 0.32 |
ENSDART00000147037
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr16_-_45917683 | 0.32 |
ENSDART00000184289
|
afp4
|
antifreeze protein type IV |
| chr15_-_35410860 | 0.32 |
ENSDART00000191267
|
mecom
|
MDS1 and EVI1 complex locus |
| chr10_+_9601587 | 0.32 |
ENSDART00000147997
|
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr4_+_30776883 | 0.32 |
ENSDART00000169519
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr23_+_19655301 | 0.32 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr14_+_909769 | 0.30 |
ENSDART00000021346
ENSDART00000172777 |
arl3l2
|
ADP-ribosylation factor-like 3, like 2 |
| chr17_-_15188440 | 0.29 |
ENSDART00000151885
|
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr7_+_15659280 | 0.29 |
ENSDART00000173414
|
mef2ab
|
myocyte enhancer factor 2ab |
| chr24_-_14712427 | 0.29 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr5_-_28767573 | 0.29 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr17_+_8324345 | 0.29 |
ENSDART00000172443
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr24_+_26328787 | 0.29 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr21_-_4695583 | 0.29 |
ENSDART00000031425
|
zgc:55582
|
zgc:55582 |
| chr23_-_35483163 | 0.29 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr22_-_36519590 | 0.28 |
ENSDART00000129318
|
CABZ01045212.1
|
|
| chr14_-_38889311 | 0.28 |
ENSDART00000186978
|
zgc:101583
|
zgc:101583 |
| chr15_-_33933790 | 0.28 |
ENSDART00000165162
ENSDART00000182258 ENSDART00000183240 |
mag
|
myelin associated glycoprotein |
| chr4_+_42450386 | 0.28 |
ENSDART00000168211
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr9_-_18911608 | 0.28 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr4_+_288633 | 0.28 |
ENSDART00000183304
|
FO834823.1
|
|
| chr5_+_28797771 | 0.28 |
ENSDART00000188845
ENSDART00000149066 |
si:ch211-186e20.7
|
si:ch211-186e20.7 |
| chr25_+_4760489 | 0.28 |
ENSDART00000167399
|
FP245455.1
|
|
| chr6_+_24420523 | 0.28 |
ENSDART00000185461
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr23_-_33709964 | 0.27 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr6_+_7322587 | 0.27 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr11_-_41853874 | 0.27 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr23_+_43950674 | 0.27 |
ENSDART00000167813
|
corin
|
corin, serine peptidase |
| chr6_-_32411703 | 0.27 |
ENSDART00000151002
ENSDART00000078908 |
usp1
|
ubiquitin specific peptidase 1 |
| chr5_-_67878064 | 0.26 |
ENSDART00000111203
|
tagln3a
|
transgelin 3a |
| chr9_+_38158570 | 0.26 |
ENSDART00000059549
ENSDART00000133060 |
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr7_+_20876303 | 0.26 |
ENSDART00000173495
ENSDART00000164172 |
gigyf1a
|
GRB10 interacting GYF protein 1a |
| chr24_-_28419444 | 0.26 |
ENSDART00000105749
|
nrros
|
negative regulator of reactive oxygen species |
| chr1_+_53714734 | 0.26 |
ENSDART00000114689
|
pus10
|
pseudouridylate synthase 10 |
| chr13_-_9895564 | 0.26 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr12_+_16132612 | 0.26 |
ENSDART00000152550
|
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr11_+_13041341 | 0.26 |
ENSDART00000166126
ENSDART00000162074 ENSDART00000170889 |
btf3l4
|
basic transcription factor 3-like 4 |
| chr11_-_45140227 | 0.26 |
ENSDART00000184062
|
cant1b
|
calcium activated nucleotidase 1b |
| chr1_-_25438737 | 0.26 |
ENSDART00000134470
|
fhdc1
|
FH2 domain containing 1 |
| chr4_-_57001458 | 0.25 |
ENSDART00000158660
|
si:ch211-161m3.1
|
si:ch211-161m3.1 |
| chr5_+_37837245 | 0.25 |
ENSDART00000171617
|
epd
|
ependymin |
| chr14_-_2004291 | 0.25 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr1_-_25438934 | 0.25 |
ENSDART00000111686
|
fhdc1
|
FH2 domain containing 1 |
| chr18_+_3634652 | 0.25 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr1_-_51219965 | 0.25 |
ENSDART00000146612
|
esf1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr4_-_9637398 | 0.24 |
ENSDART00000150253
|
dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr11_-_3535537 | 0.24 |
ENSDART00000165329
ENSDART00000009788 |
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr2_+_1714640 | 0.24 |
ENSDART00000086761
ENSDART00000111613 |
adgrl2b.1
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr10_+_15024772 | 0.24 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr9_-_2572790 | 0.24 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr15_+_25156382 | 0.24 |
ENSDART00000171092
|
slc35f2l
|
info solute carrier family 35, member F2, like |
| chr25_-_8138122 | 0.24 |
ENSDART00000104659
|
sergef
|
secretion regulating guanine nucleotide exchange factor |
| chr24_-_3783258 | 0.23 |
ENSDART00000139596
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr8_+_53423408 | 0.23 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr9_-_41040492 | 0.23 |
ENSDART00000163164
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr16_-_28091597 | 0.23 |
ENSDART00000166595
|
CR626941.1
|
|
| chr6_+_58698475 | 0.23 |
ENSDART00000056138
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr9_+_34232503 | 0.23 |
ENSDART00000132836
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr17_+_47116500 | 0.23 |
ENSDART00000186627
|
CLIP4
|
si:dkeyp-47f9.4 |
| chr20_-_26537171 | 0.23 |
ENSDART00000061926
|
stx11b.2
|
syntaxin 11b, tandem duplicate 2 |
| chr17_-_5424175 | 0.23 |
ENSDART00000171063
ENSDART00000188102 |
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr25_+_34984333 | 0.23 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr15_+_44201056 | 0.23 |
ENSDART00000162433
ENSDART00000148336 |
CU655961.4
|
|
| chr25_-_7670616 | 0.23 |
ENSDART00000131583
ENSDART00000142794 |
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr24_-_3783497 | 0.22 |
ENSDART00000158354
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr11_+_1628538 | 0.22 |
ENSDART00000154967
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr19_-_47323267 | 0.22 |
ENSDART00000190077
|
CU138512.1
|
|
| chr9_+_13229585 | 0.22 |
ENSDART00000154879
ENSDART00000141705 |
carf
|
calcium responsive transcription factor |
| chr12_+_16168342 | 0.22 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr21_+_43702016 | 0.22 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr22_-_2959005 | 0.22 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr8_+_7975745 | 0.22 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr14_+_22114918 | 0.22 |
ENSDART00000166610
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr13_+_40770628 | 0.22 |
ENSDART00000085846
|
nkx1.2la
|
NK1 transcription factor related 2-like,a |
| chr15_+_31276869 | 0.21 |
ENSDART00000156355
|
si:ch211-244e12.7
|
si:ch211-244e12.7 |
| chr11_+_3585934 | 0.21 |
ENSDART00000055694
|
cdab
|
cytidine deaminase b |
| chr16_-_41488023 | 0.21 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr6_-_11091449 | 0.21 |
ENSDART00000127209
|
pttg1ipa
|
PTTG1 interacting protein a |
| chr18_-_2433011 | 0.21 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr9_+_24708653 | 0.21 |
ENSDART00000065136
|
CT030712.1
|
|
| chr10_+_33573838 | 0.21 |
ENSDART00000051197
ENSDART00000130093 |
c10h21orf59
|
c10h21orf59 homolog (H. sapiens) |
| chr7_-_30779575 | 0.21 |
ENSDART00000004782
|
mphosph10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr23_-_16843649 | 0.21 |
ENSDART00000129971
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr7_+_20917966 | 0.21 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr10_+_16092671 | 0.21 |
ENSDART00000182761
ENSDART00000154835 |
megf10
|
multiple EGF-like-domains 10 |
| chr7_-_30926030 | 0.20 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr21_+_41468210 | 0.20 |
ENSDART00000192287
|
bod1
|
biorientation of chromosomes in cell division 1 |
| chr21_-_5077715 | 0.20 |
ENSDART00000081954
|
haus1
|
HAUS augmin-like complex, subunit 1 |
| chr2_+_42871831 | 0.20 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr15_-_26844591 | 0.20 |
ENSDART00000077582
|
pitpnm3
|
PITPNM family member 3 |
| chr7_-_5191797 | 0.20 |
ENSDART00000172955
|
si:ch73-223f5.1
|
si:ch73-223f5.1 |
| chr8_+_41229233 | 0.20 |
ENSDART00000131135
|
zgc:152830
|
zgc:152830 |
| chr16_+_5184402 | 0.20 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr25_-_22889519 | 0.20 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr15_-_2519640 | 0.20 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr5_+_45990046 | 0.20 |
ENSDART00000084024
|
sv2c
|
synaptic vesicle glycoprotein 2C |
| chr6_+_3680651 | 0.20 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| chr18_+_40471826 | 0.20 |
ENSDART00000098806
|
ugt5c3
|
UDP glucuronosyltransferase 5 family, polypeptide C3 |
| chr25_+_31323978 | 0.19 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr2_-_32825917 | 0.19 |
ENSDART00000180409
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr6_-_16406210 | 0.19 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr5_-_33886495 | 0.19 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr21_+_21692307 | 0.19 |
ENSDART00000139330
|
or125-3
|
odorant receptor, family E, subfamily 125, member 3 |
| chr15_-_21165237 | 0.19 |
ENSDART00000157069
|
A2ML1 (1 of many)
|
si:ch211-212c13.8 |
| chr13_-_18695289 | 0.19 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr6_-_55399214 | 0.19 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr25_-_29074064 | 0.18 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr11_+_36408636 | 0.18 |
ENSDART00000182098
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr25_-_3132060 | 0.18 |
ENSDART00000014363
|
epx
|
eosinophil peroxidase |
| chr15_-_1765098 | 0.18 |
ENSDART00000149980
ENSDART00000093074 |
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr21_-_3825135 | 0.18 |
ENSDART00000133792
|
st6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr20_+_54037138 | 0.18 |
ENSDART00000143172
|
wdr20b
|
WD repeat domain 20b |
| chr3_-_61362398 | 0.18 |
ENSDART00000156177
|
si:dkey-111k8.3
|
si:dkey-111k8.3 |
| chr9_+_33207574 | 0.18 |
ENSDART00000055897
ENSDART00000166030 |
si:ch211-125e6.11
|
si:ch211-125e6.11 |
| chr17_+_24632440 | 0.18 |
ENSDART00000157092
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr9_+_32301017 | 0.18 |
ENSDART00000127916
ENSDART00000183298 ENSDART00000143103 |
hspe1
|
heat shock 10 protein 1 |
| chr17_-_37184655 | 0.18 |
ENSDART00000180447
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr18_-_24988645 | 0.18 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr14_-_35414559 | 0.18 |
ENSDART00000145033
|
rnaseh2c
|
ribonuclease H2, subunit C |
| chr23_-_35790235 | 0.18 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr10_-_41940302 | 0.18 |
ENSDART00000033121
|
morn3
|
MORN repeat containing 3 |
| chr22_-_11623063 | 0.18 |
ENSDART00000145653
|
gcga
|
glucagon a |
| chr24_+_13925066 | 0.18 |
ENSDART00000134221
ENSDART00000012253 ENSDART00000081595 ENSDART00000136443 |
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr10_-_25204034 | 0.17 |
ENSDART00000161100
|
FAT3 (1 of many)
|
si:ch211-214k5.6 |
| chr2_+_40294313 | 0.17 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr6_-_57635577 | 0.17 |
ENSDART00000168708
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr16_-_8927425 | 0.17 |
ENSDART00000000382
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr10_+_40160502 | 0.17 |
ENSDART00000171879
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr13_-_40316367 | 0.17 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr10_-_11012000 | 0.17 |
ENSDART00000132995
|
ak3
|
adenylate kinase 3 |
| chr22_+_19247255 | 0.17 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr14_-_22015232 | 0.17 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr16_-_21620947 | 0.17 |
ENSDART00000115011
ENSDART00000183125 ENSDART00000188856 ENSDART00000189460 |
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr8_-_4327473 | 0.17 |
ENSDART00000134378
|
cux2b
|
cut-like homeobox 2b |
| chr21_-_21530868 | 0.17 |
ENSDART00000174231
|
or133-9
|
odorant receptor, family H, subfamily 133, member 9 |
| chr23_-_5683147 | 0.17 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr4_+_74928675 | 0.17 |
ENSDART00000192755
|
nup50
|
nucleoporin 50 |
| chr7_+_30178453 | 0.17 |
ENSDART00000174420
|
si:ch211-107o10.3
|
si:ch211-107o10.3 |
| chr3_-_46410387 | 0.17 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr24_-_25166416 | 0.17 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr4_+_17671164 | 0.17 |
ENSDART00000040445
|
gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr18_+_20468157 | 0.17 |
ENSDART00000100665
ENSDART00000147867 ENSDART00000060302 ENSDART00000180370 |
ddb2
|
damage-specific DNA binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of arid5a+arid6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 0.5 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.4 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 0.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.3 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.1 | 0.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.3 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.1 | 0.4 | GO:0070814 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.2 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.1 | 0.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.2 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.6 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.1 | 0.5 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 0.2 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.1 | 0.3 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 | 0.3 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 0.2 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.5 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.3 | GO:0071632 | optomotor response(GO:0071632) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.4 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.0 | 0.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.1 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.2 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.3 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.2 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.6 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.1 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.4 | GO:0098754 | detoxification(GO:0098754) |
| 0.0 | 0.2 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.2 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.1 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.0 | 0.0 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.0 | GO:0003218 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.0 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:1990077 | primosome complex(GO:1990077) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0030681 | fibrillar center(GO:0001650) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.3 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.4 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.2 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.1 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.6 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.0 | 0.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.3 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.1 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 1.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |