Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
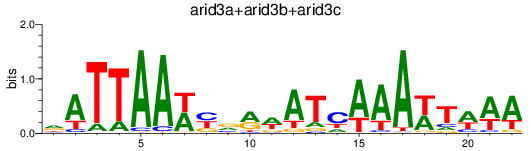
Results for arid3a+arid3b+arid3c
Z-value: 1.55
Transcription factors associated with arid3a+arid3b+arid3c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid3c
|
ENSDARG00000067729 | AT rich interactive domain 3C (BRIGHT-like) |
|
arid3a
|
ENSDARG00000070843 | AT rich interactive domain 3A (BRIGHT-like) |
|
arid3b
|
ENSDARG00000104034 | AT rich interactive domain 3B (BRIGHT-like) |
|
arid3c
|
ENSDARG00000114447 | AT rich interactive domain 3C (BRIGHT-like) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid3a | dr11_v1_chr11_+_5681762_5681762 | 0.92 | 1.6e-08 | Click! |
| arid3c | dr11_v1_chr5_+_41322783_41322783 | 0.68 | 1.5e-03 | Click! |
| arid3b | dr11_v1_chr25_-_29072162_29072162 | 0.39 | 1.0e-01 | Click! |
Activity profile of arid3a+arid3b+arid3c motif
Sorted Z-values of arid3a+arid3b+arid3c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_21225750 | 9.37 |
ENSDART00000139213
|
zgc:153968
|
zgc:153968 |
| chr19_+_4139065 | 9.18 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr18_-_47662696 | 5.58 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr2_-_9489611 | 5.44 |
ENSDART00000146715
|
st6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr10_+_23537576 | 5.38 |
ENSDART00000130515
|
CR847844.1
|
|
| chr8_+_3820134 | 5.18 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr9_+_1139378 | 4.82 |
ENSDART00000170033
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr23_+_21544227 | 4.21 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr19_-_11208782 | 4.18 |
ENSDART00000044426
ENSDART00000189754 |
si:dkey-240h12.4
|
si:dkey-240h12.4 |
| chr5_+_5398966 | 3.86 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr10_+_45031398 | 3.68 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr9_+_1137430 | 2.95 |
ENSDART00000187062
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr22_+_7439186 | 2.59 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr2_-_1548330 | 2.59 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr2_+_31492662 | 2.57 |
ENSDART00000123495
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr9_+_2574122 | 2.51 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr20_+_52389858 | 2.48 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr21_+_11401247 | 2.30 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr4_+_14660769 | 2.20 |
ENSDART00000168152
ENSDART00000013990 ENSDART00000079987 |
abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr2_-_39558643 | 2.16 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr2_+_49232559 | 2.03 |
ENSDART00000154285
ENSDART00000175147 |
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr12_-_30338779 | 2.03 |
ENSDART00000192511
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr10_+_45089820 | 2.01 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr19_-_3167729 | 1.90 |
ENSDART00000110763
ENSDART00000145710 ENSDART00000074620 ENSDART00000105174 |
stm
|
starmaker |
| chr12_+_2428247 | 1.88 |
ENSDART00000152529
|
lrrc18b
|
leucine rich repeat containing 18b |
| chr8_-_29822527 | 1.81 |
ENSDART00000167487
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr19_-_40191358 | 1.77 |
ENSDART00000183919
|
grn1
|
granulin 1 |
| chr18_+_11858397 | 1.76 |
ENSDART00000133762
|
tmtc2b
|
transmembrane and tetratricopeptide repeat containing 2b |
| chr15_-_35410860 | 1.68 |
ENSDART00000191267
|
mecom
|
MDS1 and EVI1 complex locus |
| chr10_-_2522588 | 1.68 |
ENSDART00000081926
|
CU856539.1
|
|
| chr14_-_7045009 | 1.66 |
ENSDART00000112082
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr19_-_26869103 | 1.64 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr7_+_9189547 | 1.62 |
ENSDART00000169783
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr25_+_37126921 | 1.61 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr25_+_14165447 | 1.60 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr15_+_47362728 | 1.60 |
ENSDART00000180712
|
CABZ01099833.1
|
|
| chr14_-_1635296 | 1.59 |
ENSDART00000186658
|
LO018564.1
|
|
| chr9_+_50316921 | 1.54 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr15_+_45994123 | 1.54 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr5_+_4436405 | 1.51 |
ENSDART00000167969
|
CABZ01079241.1
|
|
| chr15_-_20190052 | 1.48 |
ENSDART00000157149
|
exoc3l2b
|
exocyst complex component 3-like 2b |
| chr3_-_5228841 | 1.42 |
ENSDART00000092373
ENSDART00000182438 |
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr12_-_13650344 | 1.40 |
ENSDART00000124364
ENSDART00000124638 ENSDART00000171929 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr4_-_74892355 | 1.39 |
ENSDART00000188725
|
PHF21B
|
PHD finger protein 21B |
| chr15_-_17813680 | 1.39 |
ENSDART00000158556
|
CT573342.2
|
|
| chr20_-_45708962 | 1.39 |
ENSDART00000124283
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr5_+_45677781 | 1.33 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr21_+_36623162 | 1.33 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr1_-_2449395 | 1.31 |
ENSDART00000103785
|
ggact.3
|
gamma-glutamylamine cyclotransferase, tandem duplicate 3 |
| chr21_-_42831033 | 1.29 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr10_-_44981295 | 1.29 |
ENSDART00000166528
|
purbb
|
purine-rich element binding protein Bb |
| chr17_+_22311413 | 1.26 |
ENSDART00000151929
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr10_+_15967643 | 1.25 |
ENSDART00000136709
|
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr3_-_55537096 | 1.25 |
ENSDART00000123544
ENSDART00000188752 |
tex2
|
testis expressed 2 |
| chr21_+_45841731 | 1.24 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr5_+_58492699 | 1.23 |
ENSDART00000181584
|
FQ378016.1
|
|
| chr11_+_24819624 | 1.22 |
ENSDART00000155514
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr5_-_68058168 | 1.20 |
ENSDART00000177026
|
rnf167
|
ring finger protein 167 |
| chr1_+_51903413 | 1.19 |
ENSDART00000135861
|
nfixa
|
nuclear factor I/Xa |
| chr15_-_34408777 | 1.19 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr6_+_58698475 | 1.19 |
ENSDART00000056138
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr20_+_46741074 | 1.18 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr5_-_67349916 | 1.16 |
ENSDART00000144092
|
mlxip
|
MLX interacting protein |
| chr11_-_43200994 | 1.16 |
ENSDART00000164700
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr4_+_15944245 | 1.15 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr10_+_16911951 | 1.15 |
ENSDART00000164933
|
UNC13B (1 of many)
|
unc-13 homolog B |
| chr1_+_52662203 | 1.14 |
ENSDART00000141530
|
osbp
|
oxysterol binding protein |
| chr10_-_4375190 | 1.14 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr9_+_22011912 | 1.13 |
ENSDART00000101897
|
crygm1
|
crystallin, gamma M1 |
| chr14_+_15620640 | 1.13 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr1_+_10771457 | 1.12 |
ENSDART00000027050
ENSDART00000137185 |
cnga3b
|
cyclic nucleotide gated channel alpha 3b |
| chr18_+_6126506 | 1.11 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr17_-_4245902 | 1.10 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr8_+_7144066 | 1.08 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr12_+_47698356 | 1.08 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr23_-_41965557 | 1.07 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr16_+_22345513 | 1.07 |
ENSDART00000078000
|
zgc:123238
|
zgc:123238 |
| chr14_+_28281744 | 1.06 |
ENSDART00000173292
|
mid2
|
midline 2 |
| chr12_-_18961289 | 1.05 |
ENSDART00000168405
|
ep300a
|
E1A binding protein p300 a |
| chr4_-_2440175 | 1.01 |
ENSDART00000058059
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr8_-_29930821 | 1.01 |
ENSDART00000125173
|
ercc6l2
|
excision repair cross-complementation group 6-like 2 |
| chr1_-_12161148 | 0.98 |
ENSDART00000166429
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr5_-_3684402 | 0.97 |
ENSDART00000184354
|
FO704648.1
|
|
| chr22_+_38049130 | 0.97 |
ENSDART00000097533
|
wwtr1
|
WW domain containing transcription regulator 1 |
| chr16_-_8927425 | 0.95 |
ENSDART00000000382
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr5_+_66250856 | 0.94 |
ENSDART00000132789
|
secisbp2
|
SECIS binding protein 2 |
| chr15_-_23467750 | 0.94 |
ENSDART00000148804
|
pdzd3a
|
PDZ domain containing 3a |
| chr7_+_8324506 | 0.94 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr8_+_9715010 | 0.94 |
ENSDART00000139414
|
gripap1
|
GRIP1 associated protein 1 |
| chr20_+_18943406 | 0.94 |
ENSDART00000193590
|
mtmr9
|
myotubularin related protein 9 |
| chr3_+_60828813 | 0.93 |
ENSDART00000128260
|
CABZ01087514.1
|
|
| chr16_+_5408748 | 0.93 |
ENSDART00000160008
ENSDART00000014024 |
plecb
|
plectin b |
| chr7_+_1530024 | 0.92 |
ENSDART00000163082
|
tox4b
|
TOX high mobility group box family member 4 b |
| chr9_-_32604414 | 0.91 |
ENSDART00000088876
ENSDART00000166502 |
satb2
|
SATB homeobox 2 |
| chr20_-_30370884 | 0.91 |
ENSDART00000062429
|
allc
|
allantoicase |
| chr6_-_10780698 | 0.91 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr15_-_40267485 | 0.89 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr5_-_12407194 | 0.89 |
ENSDART00000125291
|
ksr2
|
kinase suppressor of ras 2 |
| chr16_-_49781481 | 0.89 |
ENSDART00000127513
|
znf385d
|
zinc finger protein 385D |
| chr12_+_8168272 | 0.88 |
ENSDART00000054092
|
arid5b
|
AT-rich interaction domain 5B |
| chr6_-_55309190 | 0.87 |
ENSDART00000162117
|
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr19_+_40856534 | 0.84 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr14_-_36763302 | 0.82 |
ENSDART00000074786
|
ctso
|
cathepsin O |
| chr14_-_962171 | 0.81 |
ENSDART00000010773
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr8_-_13823091 | 0.81 |
ENSDART00000177174
ENSDART00000137021 |
cabp4
|
calcium binding protein 4 |
| chr3_+_26019426 | 0.80 |
ENSDART00000135389
ENSDART00000182411 |
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr11_-_891674 | 0.79 |
ENSDART00000172904
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr2_+_52847049 | 0.77 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr25_+_34749187 | 0.77 |
ENSDART00000141473
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr11_-_31276064 | 0.77 |
ENSDART00000141062
ENSDART00000004780 |
man2b1
|
mannosidase, alpha, class 2B, member 1 |
| chr8_+_25145464 | 0.76 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr5_-_25576462 | 0.75 |
ENSDART00000165147
|
si:dkey-229d2.4
|
si:dkey-229d2.4 |
| chr19_+_32401278 | 0.75 |
ENSDART00000184353
|
atxn1a
|
ataxin 1a |
| chr13_-_41546779 | 0.74 |
ENSDART00000163331
|
pcdh15a
|
protocadherin-related 15a |
| chr19_-_47323267 | 0.74 |
ENSDART00000190077
|
CU138512.1
|
|
| chr15_-_2754056 | 0.74 |
ENSDART00000129380
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr16_-_4610255 | 0.74 |
ENSDART00000081852
ENSDART00000123253 ENSDART00000127554 ENSDART00000029485 |
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr8_+_48965767 | 0.73 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr15_+_34592215 | 0.71 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr3_-_29671653 | 0.71 |
ENSDART00000132083
|
sult3st1
|
sulfotransferase family 3, cytosolic sulfotransferase 1 |
| chr25_+_34407529 | 0.70 |
ENSDART00000156751
|
OTUD7A
|
si:dkey-37f18.2 |
| chr21_-_2707768 | 0.70 |
ENSDART00000165384
|
CABZ01101739.1
|
|
| chr10_+_22510280 | 0.69 |
ENSDART00000109070
ENSDART00000182002 ENSDART00000192852 |
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr9_+_34641237 | 0.69 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr10_-_14943281 | 0.69 |
ENSDART00000143608
|
smad2
|
SMAD family member 2 |
| chr5_-_54197084 | 0.68 |
ENSDART00000163640
|
grk1b
|
G protein-coupled receptor kinase 1 b |
| chr22_-_15562933 | 0.68 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr7_-_8324927 | 0.68 |
ENSDART00000102535
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr8_+_20950401 | 0.68 |
ENSDART00000136484
ENSDART00000131329 ENSDART00000144451 ENSDART00000144929 |
si:dkeyp-82a1.1
|
si:dkeyp-82a1.1 |
| chr7_+_60551133 | 0.67 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr23_-_32100106 | 0.66 |
ENSDART00000044658
|
letmd1
|
LETM1 domain containing 1 |
| chr4_-_20027571 | 0.65 |
ENSDART00000003420
|
drd4-rs
|
dopamine receptor D4 related sequence |
| chr3_+_7808459 | 0.65 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr9_+_33158191 | 0.65 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr18_-_19103929 | 0.65 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr19_+_43753995 | 0.65 |
ENSDART00000058504
|
SLC17A3
|
si:ch1073-513e17.1 |
| chr23_+_2666944 | 0.65 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr15_-_1733235 | 0.65 |
ENSDART00000023153
ENSDART00000154668 |
rabgef1l
|
RAB guanine nucleotide exchange factor (GEF) 1, like |
| chr4_-_63166758 | 0.64 |
ENSDART00000192765
|
CR450780.1
|
|
| chr11_-_1400507 | 0.62 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr2_+_3472832 | 0.61 |
ENSDART00000115278
|
cx47.1
|
connexin 47.1 |
| chr10_+_2669405 | 0.61 |
ENSDART00000110202
|
ccl27b
|
chemokine (C-C motif) ligand 27b |
| chr4_+_31405646 | 0.61 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr9_-_40765868 | 0.61 |
ENSDART00000138634
|
abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr22_+_30047245 | 0.60 |
ENSDART00000142857
ENSDART00000141247 ENSDART00000140015 ENSDART00000040538 |
add3a
|
adducin 3 (gamma) a |
| chr5_-_30620625 | 0.60 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr6_-_7052408 | 0.59 |
ENSDART00000150033
ENSDART00000149232 |
bin1b
|
bridging integrator 1b |
| chr19_+_47299212 | 0.59 |
ENSDART00000158262
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr2_+_30368800 | 0.59 |
ENSDART00000179564
|
pi15b
|
peptidase inhibitor 15b |
| chr6_-_9563534 | 0.59 |
ENSDART00000184765
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr8_+_43340995 | 0.59 |
ENSDART00000038566
|
rflna
|
refilin A |
| chr22_-_10891213 | 0.59 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr4_+_75292781 | 0.58 |
ENSDART00000082522
|
abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr13_-_5978433 | 0.58 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr3_-_21280373 | 0.57 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr15_-_31109760 | 0.57 |
ENSDART00000154254
|
ksr1b
|
kinase suppressor of ras 1b |
| chr1_+_2321384 | 0.57 |
ENSDART00000157662
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr3_-_21037840 | 0.57 |
ENSDART00000002393
|
rundc3aa
|
RUN domain containing 3Aa |
| chr5_-_13251907 | 0.56 |
ENSDART00000176774
ENSDART00000030553 |
top3b
|
DNA topoisomerase III beta |
| chr7_-_7692992 | 0.56 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr20_-_630637 | 0.55 |
ENSDART00000160454
|
si:dkey-121j17.5
|
si:dkey-121j17.5 |
| chr13_+_25199849 | 0.54 |
ENSDART00000139209
ENSDART00000130876 |
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr25_+_34407740 | 0.53 |
ENSDART00000012677
|
OTUD7A
|
si:dkey-37f18.2 |
| chr17_+_8292892 | 0.53 |
ENSDART00000125728
|
si:ch211-236p5.3
|
si:ch211-236p5.3 |
| chr14_+_45028062 | 0.53 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr11_+_2687395 | 0.53 |
ENSDART00000082510
|
b3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr23_+_4777348 | 0.52 |
ENSDART00000171971
|
vgll4a
|
vestigial-like family member 4a |
| chr12_-_34758474 | 0.52 |
ENSDART00000153418
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr8_+_1651821 | 0.52 |
ENSDART00000060865
ENSDART00000186304 |
rasal1b
|
RAS protein activator like 1b (GAP1 like) |
| chr12_-_33659328 | 0.52 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr19_-_40199081 | 0.51 |
ENSDART00000051970
ENSDART00000151079 |
grn2
|
granulin 2 |
| chr23_+_9035578 | 0.51 |
ENSDART00000140455
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr16_+_17913936 | 0.51 |
ENSDART00000162464
ENSDART00000149097 |
rorc
|
RAR-related orphan receptor C |
| chr1_+_47446032 | 0.51 |
ENSDART00000126904
ENSDART00000007262 |
gja8b
|
gap junction protein alpha 8 paralog b |
| chr21_+_10794914 | 0.50 |
ENSDART00000084035
|
znf532
|
zinc finger protein 532 |
| chr6_-_1865323 | 0.50 |
ENSDART00000155775
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr24_+_37484661 | 0.49 |
ENSDART00000165125
ENSDART00000109221 |
wdr90
|
WD repeat domain 90 |
| chr1_-_9227804 | 0.49 |
ENSDART00000190360
|
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr19_+_24872159 | 0.49 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr7_+_16047863 | 0.49 |
ENSDART00000188750
|
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr16_+_1100559 | 0.49 |
ENSDART00000092657
|
adamts16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr22_+_2751887 | 0.49 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr9_+_3429662 | 0.48 |
ENSDART00000160977
ENSDART00000114168 ENSDART00000082153 |
CU469503.1
itga6a
|
integrin, alpha 6a |
| chr3_+_62339264 | 0.48 |
ENSDART00000174569
|
BX470259.3
|
|
| chr9_+_38502524 | 0.48 |
ENSDART00000087229
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr21_+_37355716 | 0.48 |
ENSDART00000131188
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr9_+_2522797 | 0.48 |
ENSDART00000186786
ENSDART00000147034 |
gpr155a
|
G protein-coupled receptor 155a |
| chr18_+_44849809 | 0.47 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr8_-_2100808 | 0.47 |
ENSDART00000123804
|
cltcl1
|
clathrin, heavy chain-like 1 |
| chr1_-_1627487 | 0.47 |
ENSDART00000166094
|
clic6
|
chloride intracellular channel 6 |
| chr8_+_31716872 | 0.47 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr1_-_51219965 | 0.46 |
ENSDART00000146612
|
esf1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr8_+_104114 | 0.46 |
ENSDART00000172101
|
sncaip
|
synuclein, alpha interacting protein |
| chr10_-_20706270 | 0.46 |
ENSDART00000166540
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr4_+_25912308 | 0.45 |
ENSDART00000167845
ENSDART00000136927 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr7_-_11596450 | 0.45 |
ENSDART00000173863
|
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr23_+_30967686 | 0.45 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr19_-_14155781 | 0.45 |
ENSDART00000169232
|
nr0b2b
|
nuclear receptor subfamily 0, group B, member 2b |
| chr14_+_30340251 | 0.45 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr3_+_24511959 | 0.45 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
Network of associatons between targets according to the STRING database.
First level regulatory network of arid3a+arid3b+arid3c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.8 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.6 | 3.9 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.6 | 1.7 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.4 | 5.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 2.6 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.3 | 4.2 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.3 | 0.9 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 0.9 | GO:0000256 | allantoin catabolic process(GO:0000256) |
| 0.3 | 1.9 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.2 | 1.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 1.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.2 | 0.9 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.2 | 1.4 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 0.9 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.2 | 2.3 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 0.6 | GO:1903011 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.2 | 1.0 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.2 | 1.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.5 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 1.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.4 | GO:0015808 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.1 | 1.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.7 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.6 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.6 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.1 | 0.9 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 5.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 3.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.4 | GO:2000561 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 1.5 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.6 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 0.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.4 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.6 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.2 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.1 | 1.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.7 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.9 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.3 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 2.0 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 1.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 1.3 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 3.3 | GO:0030203 | glycosaminoglycan metabolic process(GO:0030203) |
| 0.0 | 1.6 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.7 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.9 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.7 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.8 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 1.2 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.5 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.6 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 2.3 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.4 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.1 | GO:0044108 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.0 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 1.6 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.6 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.4 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 3.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.9 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 3.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 0.9 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.4 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.1 | 1.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 8.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 2.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 5.2 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.3 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 4.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.8 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.8 | 2.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.7 | 5.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.5 | 1.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.4 | 1.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.4 | 2.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.4 | 1.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.3 | 4.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 0.9 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.2 | 3.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.6 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.2 | 1.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 0.9 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 0.9 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.2 | 1.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.7 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 1.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.8 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.5 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 1.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.1 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 2.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.4 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.4 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 1.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.8 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 3.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.4 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.9 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 2.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 7.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 2.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.5 | GO:0017171 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 1.6 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 6.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 1.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 4.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 1.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 2.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 3.9 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 3.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 0.8 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |