Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
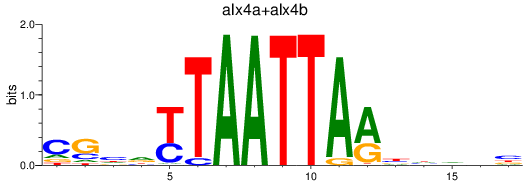
Results for alx4a+alx4b
Z-value: 0.36
Transcription factors associated with alx4a+alx4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
alx4b
|
ENSDARG00000074442 | ALX homeobox 4b |
|
alx4a
|
ENSDARG00000088332 | ALX homeobox 4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| alx4a | dr11_v1_chr7_-_26924903_26924903 | -0.24 | 3.2e-01 | Click! |
| alx4b | dr11_v1_chr18_+_38321039_38321039 | 0.22 | 3.6e-01 | Click! |
Activity profile of alx4a+alx4b motif
Sorted Z-values of alx4a+alx4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22099536 | 1.85 |
ENSDART00000101923
|
CR391987.1
|
|
| chr22_+_7738966 | 1.26 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr17_-_37395460 | 1.11 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr3_-_31079186 | 1.07 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr25_+_29161609 | 0.94 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr9_-_22129788 | 0.85 |
ENSDART00000124272
ENSDART00000175417 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr15_+_45640906 | 0.80 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr1_-_43915423 | 0.78 |
ENSDART00000181915
ENSDART00000113673 |
scpp5
|
secretory calcium-binding phosphoprotein 5 |
| chr3_+_24361096 | 0.74 |
ENSDART00000132387
|
pvalb6
|
parvalbumin 6 |
| chr8_+_34731982 | 0.72 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr6_-_607063 | 0.66 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr2_-_38000276 | 0.66 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr16_-_11798994 | 0.65 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr17_-_12336987 | 0.61 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr9_+_2574122 | 0.58 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr14_-_49063157 | 0.58 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr24_-_7697274 | 0.56 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr16_-_28658341 | 0.56 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr21_+_21195487 | 0.54 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr16_-_12173554 | 0.54 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr5_+_2815021 | 0.52 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr25_+_10811551 | 0.51 |
ENSDART00000167730
|
anpepb
|
alanyl (membrane) aminopeptidase b |
| chr15_-_5815006 | 0.50 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr3_+_62000822 | 0.49 |
ENSDART00000106680
|
rai1
|
retinoic acid induced 1 |
| chr25_+_31276842 | 0.47 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr5_-_23280098 | 0.44 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr25_+_5821910 | 0.43 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr25_+_14087045 | 0.42 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr1_-_50438247 | 0.42 |
ENSDART00000114098
|
dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr7_-_73752955 | 0.42 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr7_-_66868543 | 0.41 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr21_-_20939488 | 0.41 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr2_+_50608099 | 0.41 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr13_-_22862133 | 0.41 |
ENSDART00000138563
|
pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr5_-_41494831 | 0.41 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr2_-_27329667 | 0.40 |
ENSDART00000187490
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr11_+_42474694 | 0.40 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr18_-_48983690 | 0.40 |
ENSDART00000182359
|
FO681288.3
|
|
| chr15_-_12258851 | 0.39 |
ENSDART00000180656
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr18_+_21408794 | 0.39 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr16_+_2820340 | 0.39 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr21_+_31150438 | 0.39 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr19_-_26869103 | 0.38 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr8_+_43053519 | 0.38 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr16_-_12173399 | 0.37 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr5_+_37068223 | 0.37 |
ENSDART00000164279
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr9_+_1139378 | 0.37 |
ENSDART00000170033
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr6_+_39905021 | 0.37 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr17_+_15433518 | 0.37 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr1_-_43948826 | 0.36 |
ENSDART00000132038
|
si:dkey-22i16.7
|
si:dkey-22i16.7 |
| chr17_-_200316 | 0.36 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr16_-_28856112 | 0.36 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr22_-_20011476 | 0.36 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr23_-_3681026 | 0.36 |
ENSDART00000192128
ENSDART00000040086 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr21_+_28958471 | 0.35 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr20_+_18661624 | 0.35 |
ENSDART00000152136
ENSDART00000126959 |
tnfaip2a
|
tumor necrosis factor, alpha-induced protein 2a |
| chr25_+_8356707 | 0.35 |
ENSDART00000153708
|
muc5.1
|
mucin 5.1, oligomeric mucus/gel-forming |
| chr12_-_26153101 | 0.34 |
ENSDART00000076051
|
opn4b
|
opsin 4b |
| chr21_+_45841731 | 0.34 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr4_+_21129752 | 0.34 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr2_+_2223837 | 0.34 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr17_+_15433671 | 0.34 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr3_+_26813058 | 0.34 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr14_+_40874608 | 0.33 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr9_-_35334642 | 0.33 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr19_+_43297546 | 0.32 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr6_+_612594 | 0.32 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr2_+_44571200 | 0.32 |
ENSDART00000098132
|
klhl24a
|
kelch-like family member 24a |
| chr20_+_9128256 | 0.31 |
ENSDART00000163883
ENSDART00000183072 ENSDART00000187276 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr3_-_21280373 | 0.31 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr12_+_31713239 | 0.31 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr6_-_43677125 | 0.31 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr1_+_59293873 | 0.30 |
ENSDART00000168036
|
rdh8b
|
retinol dehydrogenase 8b |
| chr24_-_29586082 | 0.30 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr10_-_42519773 | 0.30 |
ENSDART00000039187
|
march5l
|
membrane-associated ring finger (C3HC4) 5, like |
| chr7_+_34620418 | 0.30 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr21_+_31150773 | 0.30 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr13_-_37229245 | 0.30 |
ENSDART00000140923
|
si:dkeyp-77c8.5
|
si:dkeyp-77c8.5 |
| chr16_-_12316979 | 0.30 |
ENSDART00000182392
|
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr22_-_10482253 | 0.29 |
ENSDART00000143164
|
aspn
|
asporin (LRR class 1) |
| chr14_-_14659023 | 0.29 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr3_+_26814030 | 0.29 |
ENSDART00000180128
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr23_-_24483311 | 0.29 |
ENSDART00000185793
ENSDART00000109248 |
spen
|
spen family transcriptional repressor |
| chr7_+_39634873 | 0.29 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr11_+_30057762 | 0.29 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr1_-_15797663 | 0.29 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr13_-_29420885 | 0.29 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr12_+_48634927 | 0.28 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr15_-_46779934 | 0.28 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr16_-_38118003 | 0.27 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr13_+_6342410 | 0.27 |
ENSDART00000065228
|
csmd1a
|
CUB and Sushi multiple domains 1a |
| chr10_+_17776981 | 0.27 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr10_-_35236949 | 0.26 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr10_-_27049170 | 0.26 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr15_-_1941042 | 0.26 |
ENSDART00000112946
ENSDART00000155504 |
dock10
|
dedicator of cytokinesis 10 |
| chr3_+_25154078 | 0.26 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr2_+_53204750 | 0.26 |
ENSDART00000163644
|
zgc:165603
|
zgc:165603 |
| chr25_-_19420949 | 0.26 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr9_+_29643036 | 0.26 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr16_+_13818500 | 0.26 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr7_+_23515966 | 0.26 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr4_+_22480169 | 0.26 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr21_-_40174647 | 0.26 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr19_+_823945 | 0.25 |
ENSDART00000142287
|
ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr9_+_54039006 | 0.25 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr14_-_4556896 | 0.25 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr15_-_12011390 | 0.25 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr11_+_44356707 | 0.25 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr15_-_33925851 | 0.25 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr15_-_462783 | 0.25 |
ENSDART00000161049
|
CABZ01033205.1
|
Danio rerio uncharacterized LOC100002960 (LOC100002960), mRNA. |
| chr6_+_24420523 | 0.25 |
ENSDART00000185461
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr1_-_49250490 | 0.25 |
ENSDART00000150386
|
si:ch73-6k14.2
|
si:ch73-6k14.2 |
| chr10_-_31015535 | 0.24 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr24_+_7800486 | 0.24 |
ENSDART00000145504
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr21_-_22635245 | 0.24 |
ENSDART00000115224
ENSDART00000101782 |
nectin1a
|
nectin cell adhesion molecule 1a |
| chr3_-_59297532 | 0.24 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr1_+_55755304 | 0.24 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr6_+_11250033 | 0.24 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr6_+_11249706 | 0.24 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr16_+_34531486 | 0.23 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr9_-_46399611 | 0.23 |
ENSDART00000164914
ENSDART00000145931 |
lct
si:dkey-79p17.3
|
lactase si:dkey-79p17.3 |
| chr19_+_5480327 | 0.23 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr11_-_2838699 | 0.23 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr14_-_858985 | 0.23 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr4_-_2219705 | 0.23 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr23_+_238893 | 0.23 |
ENSDART00000138198
|
PDZD4
|
si:ch73-162j3.4 |
| chr11_+_44356504 | 0.23 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr5_+_64739762 | 0.23 |
ENSDART00000161112
ENSDART00000135610 ENSDART00000002908 |
olfm1a
|
olfactomedin 1a |
| chr19_-_27858033 | 0.23 |
ENSDART00000103898
ENSDART00000144884 |
srd5a1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr7_+_24573721 | 0.23 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr13_-_20381485 | 0.23 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr8_-_13184989 | 0.22 |
ENSDART00000135738
|
zgc:194990
|
zgc:194990 |
| chr7_-_71829649 | 0.22 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr4_+_9669717 | 0.22 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr23_-_20051369 | 0.22 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr2_+_366429 | 0.22 |
ENSDART00000111746
|
si:dkey-33c14.6
|
si:dkey-33c14.6 |
| chr10_-_13343831 | 0.22 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr18_+_23218980 | 0.22 |
ENSDART00000185014
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr12_-_41684729 | 0.22 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr7_-_72261721 | 0.22 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr21_-_275377 | 0.22 |
ENSDART00000157509
|
rln1
|
relaxin 1 |
| chr2_-_48153945 | 0.22 |
ENSDART00000146553
|
pfkpb
|
phosphofructokinase, platelet b |
| chr12_+_18578597 | 0.22 |
ENSDART00000134944
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr19_-_15397946 | 0.21 |
ENSDART00000143480
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr6_-_39489190 | 0.21 |
ENSDART00000151299
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr21_+_8276989 | 0.21 |
ENSDART00000028265
|
nr5a1b
|
nuclear receptor subfamily 5, group A, member 1b |
| chr9_-_43538328 | 0.21 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr11_-_669558 | 0.21 |
ENSDART00000173450
|
pparg
|
peroxisome proliferator-activated receptor gamma |
| chr8_-_45838277 | 0.21 |
ENSDART00000046064
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr1_+_9966384 | 0.21 |
ENSDART00000132607
|
si:dkeyp-75b4.8
|
si:dkeyp-75b4.8 |
| chr21_+_23108420 | 0.21 |
ENSDART00000192394
ENSDART00000088459 |
htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr15_-_1885247 | 0.21 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr13_+_7292061 | 0.21 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr20_-_9436521 | 0.21 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr17_+_51627209 | 0.21 |
ENSDART00000056886
|
zgc:113142
|
zgc:113142 |
| chr2_+_55916911 | 0.21 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr6_+_25282671 | 0.21 |
ENSDART00000157575
|
si:ch211-180m24.3
|
si:ch211-180m24.3 |
| chr15_-_44601331 | 0.20 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr2_+_3986083 | 0.20 |
ENSDART00000188979
|
mkxb
|
mohawk homeobox b |
| chr3_+_28953274 | 0.20 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr19_-_5103313 | 0.20 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr19_-_20113696 | 0.20 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr15_+_34592215 | 0.19 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr5_-_16218777 | 0.19 |
ENSDART00000141698
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr17_-_36896560 | 0.19 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr8_+_7359294 | 0.19 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr12_-_41662327 | 0.19 |
ENSDART00000191602
ENSDART00000170423 |
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr23_-_16692312 | 0.19 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr3_-_32818607 | 0.19 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr11_-_2131280 | 0.19 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr8_+_52637507 | 0.19 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr20_+_591505 | 0.19 |
ENSDART00000046438
|
kcnk2b
|
potassium channel, subfamily K, member 2b |
| chr10_-_2524297 | 0.19 |
ENSDART00000192475
|
CU856539.1
|
|
| chr18_+_39487486 | 0.19 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr25_-_35136267 | 0.19 |
ENSDART00000109751
|
zgc:173585
|
zgc:173585 |
| chr5_+_37903790 | 0.19 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr22_-_12160283 | 0.19 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr20_+_3108597 | 0.19 |
ENSDART00000133435
|
CEP170B (1 of many)
|
si:ch73-212j7.1 |
| chr6_+_11250316 | 0.18 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr16_-_31622777 | 0.18 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr9_+_34127005 | 0.18 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr22_+_1556948 | 0.18 |
ENSDART00000159050
|
si:ch211-255f4.8
|
si:ch211-255f4.8 |
| chr7_-_66877058 | 0.18 |
ENSDART00000155954
|
adma
|
adrenomedullin a |
| chr3_+_33341640 | 0.18 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr7_-_71531846 | 0.18 |
ENSDART00000111797
|
acer2
|
alkaline ceramidase 2 |
| chr13_+_1089942 | 0.18 |
ENSDART00000054322
|
cnrip1b
|
cannabinoid receptor interacting protein 1b |
| chr13_-_18695289 | 0.18 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr6_+_27151940 | 0.18 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr20_+_66857 | 0.18 |
ENSDART00000114999
|
lrfn2b
|
leucine rich repeat and fibronectin type III domain containing 2b |
| chr12_-_6880694 | 0.18 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr17_+_38476300 | 0.18 |
ENSDART00000123298
|
stard9
|
StAR-related lipid transfer (START) domain containing 9 |
| chr14_-_34276574 | 0.18 |
ENSDART00000021437
|
gria1a
|
glutamate receptor, ionotropic, AMPA 1a |
| chr10_-_11261565 | 0.18 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr12_+_31735159 | 0.18 |
ENSDART00000185442
|
RNF157
|
si:dkey-49c17.3 |
| chr2_+_48073972 | 0.18 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr10_+_15255198 | 0.18 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr15_-_22074315 | 0.17 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr20_+_32552912 | 0.17 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr13_-_2010191 | 0.17 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr6_-_37468971 | 0.17 |
ENSDART00000126379
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of alx4a+alx4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.2 | 1.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.7 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.3 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.1 | 0.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.4 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.3 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.2 | GO:0044406 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.2 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.8 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.1 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.1 | 0.7 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.4 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.2 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.2 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.0 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.1 | GO:0044038 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0070169 | positive regulation of bone mineralization(GO:0030501) positive regulation of osteoblast differentiation(GO:0045669) positive regulation of ossification(GO:0045778) positive regulation of biomineral tissue development(GO:0070169) |
| 0.0 | 0.3 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.0 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.2 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.9 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.6 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.4 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:1900186 | optomotor response(GO:0071632) caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.0 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.1 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.3 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.7 | GO:0007599 | hemostasis(GO:0007599) |
| 0.0 | 0.1 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 0.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.1 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0001556 | oocyte maturation(GO:0001556) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 0.6 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 0.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.3 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.4 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.2 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.4 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.2 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.3 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.3 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |