Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
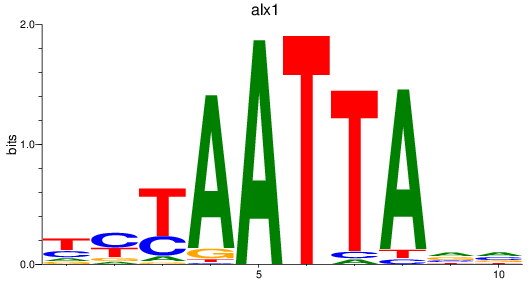
Results for alx1
Z-value: 1.65
Transcription factors associated with alx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
alx1
|
ENSDARG00000062824 | ALX homeobox 1 |
|
alx1
|
ENSDARG00000110530 | ALX homeobox 1 |
|
alx1
|
ENSDARG00000115230 | ALX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| alx1 | dr11_v1_chr18_+_16246806_16246806 | 0.62 | 5.0e-03 | Click! |
Activity profile of alx1 motif
Sorted Z-values of alx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 19.81 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr1_-_43905252 | 9.62 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr4_-_13580348 | 7.86 |
ENSDART00000067160
|
opn1sw1
|
opsin 1 (cone pigments), short-wave-sensitive 1 |
| chr3_-_31079186 | 5.85 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr10_-_11385155 | 5.55 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr20_-_40755614 | 5.38 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr19_+_10339538 | 4.98 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr5_+_51594209 | 4.95 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr25_+_31227747 | 4.62 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr2_+_50608099 | 4.57 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr2_-_51794472 | 4.47 |
ENSDART00000186652
|
BX908782.3
|
|
| chr25_+_31277415 | 4.42 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr17_-_37395460 | 4.41 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr9_+_34641237 | 4.31 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr23_-_27571667 | 4.30 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr23_+_40460333 | 4.25 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr16_+_5774977 | 4.24 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr15_+_47903864 | 4.00 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr24_-_31452875 | 3.91 |
ENSDART00000187381
ENSDART00000185128 |
cngb3.2
|
cyclic nucleotide gated channel beta 3, tandem duplicate 2 |
| chr20_-_52902693 | 3.89 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr6_-_46861676 | 3.83 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr14_-_2933185 | 3.82 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr21_+_42226113 | 3.76 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr19_-_5103141 | 3.76 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr18_-_16801033 | 3.70 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr15_-_34408777 | 3.64 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr23_+_20563779 | 3.60 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr24_-_7697274 | 3.54 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr20_+_4060839 | 3.35 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr15_-_16098531 | 3.35 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr18_+_15644559 | 3.31 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_-_23280098 | 3.25 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr1_-_50859053 | 3.23 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr21_-_14251306 | 3.19 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr4_+_12612723 | 3.18 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr13_+_29462249 | 3.12 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr16_-_28658341 | 3.10 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr21_+_10739846 | 3.03 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr23_-_39666519 | 3.03 |
ENSDART00000110868
ENSDART00000190961 |
vwa1
|
von Willebrand factor A domain containing 1 |
| chr23_+_45584223 | 3.00 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr22_-_10486477 | 2.96 |
ENSDART00000184366
|
aspn
|
asporin (LRR class 1) |
| chr5_-_41494831 | 2.95 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr7_+_71547981 | 2.85 |
ENSDART00000012070
|
adcyap1a
|
adenylate cyclase activating polypeptide 1a |
| chr6_-_50203682 | 2.80 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr7_-_24699985 | 2.73 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr14_+_45675306 | 2.68 |
ENSDART00000105461
|
rom1b
|
retinal outer segment membrane protein 1b |
| chr25_+_7504314 | 2.68 |
ENSDART00000163231
|
ifitm5
|
interferon induced transmembrane protein 5 |
| chr21_-_22737228 | 2.64 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr20_-_39271844 | 2.62 |
ENSDART00000192708
|
clu
|
clusterin |
| chr22_+_20427170 | 2.59 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr7_+_73397283 | 2.56 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr15_-_46779934 | 2.56 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr12_+_2522642 | 2.55 |
ENSDART00000152567
|
frmpd2
|
FERM and PDZ domain containing 2 |
| chr19_-_5103313 | 2.53 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr20_-_9436521 | 2.51 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr6_+_9175886 | 2.51 |
ENSDART00000165333
|
si:ch211-207l14.1
|
si:ch211-207l14.1 |
| chr8_-_14126646 | 2.48 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr2_+_20332044 | 2.48 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr23_-_19230627 | 2.43 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr6_+_9870192 | 2.42 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr4_+_21129752 | 2.39 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr10_-_7756865 | 2.39 |
ENSDART00000114373
ENSDART00000125407 ENSDART00000016317 |
loxa
|
lysyl oxidase a |
| chr17_-_16965809 | 2.38 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr10_+_29698467 | 2.35 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr1_+_8601935 | 2.34 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr20_+_18551657 | 2.33 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr16_-_45058919 | 2.30 |
ENSDART00000177134
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr21_+_13366353 | 2.28 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr8_-_46525092 | 2.28 |
ENSDART00000030482
|
sult1st2
|
sulfotransferase family 1, cytosolic sulfotransferase 2 |
| chr2_+_37227011 | 2.27 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr4_+_8797197 | 2.26 |
ENSDART00000158671
|
sult4a1
|
sulfotransferase family 4A, member 1 |
| chr7_+_71547747 | 2.26 |
ENSDART00000180869
|
adcyap1a
|
adenylate cyclase activating polypeptide 1a |
| chr10_-_27049170 | 2.25 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr25_-_12937727 | 2.22 |
ENSDART00000172643
|
ccl39.6
|
chemokine (C-C motif) ligand 39, duplicate 6 |
| chr7_+_48761875 | 2.21 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr5_+_37068223 | 2.19 |
ENSDART00000164279
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr1_-_10647484 | 2.19 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr2_+_3986083 | 2.18 |
ENSDART00000188979
|
mkxb
|
mohawk homeobox b |
| chr8_+_43053519 | 2.17 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr21_+_28958471 | 2.15 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr13_+_38430466 | 2.15 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr25_-_25384045 | 2.15 |
ENSDART00000150631
|
zgc:123278
|
zgc:123278 |
| chr25_+_3327071 | 2.13 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr9_-_14683574 | 2.12 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr21_-_22827548 | 2.11 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr7_-_27685365 | 2.11 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr25_+_33849647 | 2.09 |
ENSDART00000121449
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr21_-_18993110 | 2.08 |
ENSDART00000144086
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr21_+_39705483 | 2.06 |
ENSDART00000147718
ENSDART00000168996 |
plrdgb
|
PITP-less RdgB-like protein |
| chr24_+_5208171 | 2.05 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr16_+_31804590 | 2.05 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr3_+_25154078 | 2.02 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr25_-_27564205 | 2.02 |
ENSDART00000157319
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr1_+_31110817 | 2.02 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr11_-_19694334 | 1.99 |
ENSDART00000054735
|
SYNPR
|
si:dkey-30j16.3 |
| chr6_-_35439406 | 1.99 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr3_-_61162750 | 1.97 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr2_+_6963296 | 1.97 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr21_-_16632808 | 1.96 |
ENSDART00000172645
|
unc5da
|
unc-5 netrin receptor Da |
| chr13_+_45709380 | 1.96 |
ENSDART00000192862
|
si:ch211-62a1.3
|
si:ch211-62a1.3 |
| chr6_-_30839763 | 1.95 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr19_+_19976990 | 1.94 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr12_+_24342303 | 1.94 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr15_-_22074315 | 1.93 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr17_-_200316 | 1.92 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr6_-_38419318 | 1.91 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr21_-_41873584 | 1.90 |
ENSDART00000188089
|
endou2
|
endonuclease, polyU-specific 2 |
| chr19_-_27261102 | 1.90 |
ENSDART00000143919
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr9_-_49493305 | 1.87 |
ENSDART00000148707
ENSDART00000148561 |
xirp2b
|
xin actin binding repeat containing 2b |
| chr11_-_24191928 | 1.87 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr16_+_2820340 | 1.86 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr21_+_44857293 | 1.86 |
ENSDART00000134365
ENSDART00000168217 ENSDART00000065083 |
fstl4
|
follistatin-like 4 |
| chr14_+_25817628 | 1.85 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr1_-_59287410 | 1.84 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr16_+_39159752 | 1.82 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr2_+_28672152 | 1.81 |
ENSDART00000157410
ENSDART00000169614 |
nadsyn1
|
NAD synthetase 1 |
| chr24_-_24282427 | 1.81 |
ENSDART00000123299
|
pdha1b
|
pyruvate dehydrogenase E1 alpha 1 subunit b |
| chr20_-_45062514 | 1.80 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr5_+_37903790 | 1.80 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr15_-_6247775 | 1.80 |
ENSDART00000148350
|
dscamb
|
Down syndrome cell adhesion molecule b |
| chr18_+_14564085 | 1.79 |
ENSDART00000009363
ENSDART00000141813 ENSDART00000136120 |
si:dkey-246g23.4
|
si:dkey-246g23.4 |
| chr8_+_635704 | 1.79 |
ENSDART00000130358
|
csgalnact1b
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1b |
| chr1_+_135903 | 1.78 |
ENSDART00000124837
|
f10
|
coagulation factor X |
| chr1_+_33969015 | 1.78 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr20_-_45060241 | 1.77 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr4_-_14315855 | 1.75 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr20_+_32523576 | 1.74 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr13_+_4225173 | 1.74 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr19_-_5805923 | 1.72 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr6_+_51773873 | 1.71 |
ENSDART00000156516
|
tmem74b
|
transmembrane protein 74B |
| chr20_-_1151265 | 1.70 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr11_+_12052791 | 1.68 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr6_+_55032439 | 1.68 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr18_+_1703984 | 1.68 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr10_-_26744131 | 1.68 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr21_+_3093419 | 1.67 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr4_+_11384891 | 1.67 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr12_-_19007834 | 1.67 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr23_-_32157865 | 1.66 |
ENSDART00000000876
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr8_+_3820134 | 1.65 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr3_-_25268751 | 1.63 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr24_-_38110779 | 1.62 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr3_+_32553714 | 1.62 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr12_+_45677293 | 1.61 |
ENSDART00000152850
ENSDART00000153047 |
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr19_-_13808630 | 1.59 |
ENSDART00000166895
ENSDART00000187670 |
ctgfb
|
connective tissue growth factor b |
| chr16_+_34531486 | 1.59 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr17_-_28198099 | 1.58 |
ENSDART00000156143
|
htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr23_-_20051369 | 1.58 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr3_+_33341640 | 1.58 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr10_-_35103208 | 1.54 |
ENSDART00000192734
|
zgc:110006
|
zgc:110006 |
| chr15_+_44366556 | 1.53 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr15_-_29598679 | 1.53 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr12_+_41697664 | 1.52 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr1_+_52392511 | 1.51 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr5_-_23999777 | 1.51 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr22_-_10121880 | 1.50 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr13_+_28701233 | 1.50 |
ENSDART00000135931
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr1_-_5455498 | 1.49 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr6_+_41038757 | 1.49 |
ENSDART00000011245
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr4_-_16334362 | 1.48 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr25_+_31267268 | 1.48 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr7_-_28148310 | 1.48 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr17_-_7861219 | 1.46 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr20_+_42565049 | 1.46 |
ENSDART00000061101
|
igf2r
|
insulin-like growth factor 2 receptor |
| chr5_+_64732036 | 1.46 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr20_-_9273669 | 1.46 |
ENSDART00000175069
|
syt14b
|
synaptotagmin XIVb |
| chr21_-_25722834 | 1.46 |
ENSDART00000101208
|
abhd11
|
abhydrolase domain containing 11 |
| chr5_-_28915130 | 1.46 |
ENSDART00000078592
|
npdc1b
|
neural proliferation, differentiation and control, 1b |
| chr23_-_32162810 | 1.44 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr16_+_13883872 | 1.44 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr17_+_43032529 | 1.42 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr9_+_24159280 | 1.42 |
ENSDART00000184624
ENSDART00000178422 |
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr15_-_21877726 | 1.40 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr7_-_38792543 | 1.40 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr13_-_29420885 | 1.39 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr22_+_18477934 | 1.39 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr19_-_5865766 | 1.38 |
ENSDART00000191007
|
LO018585.1
|
|
| chr17_-_30666037 | 1.37 |
ENSDART00000156509
|
alkal2b
|
ALK and LTK ligand 2b |
| chr7_-_38658411 | 1.37 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr6_+_13730522 | 1.37 |
ENSDART00000153524
|
wnt6a
|
wingless-type MMTV integration site family, member 6a |
| chr21_-_21089781 | 1.37 |
ENSDART00000144361
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr25_-_13381854 | 1.36 |
ENSDART00000164621
ENSDART00000169129 |
ndrg4
|
NDRG family member 4 |
| chr21_-_37790727 | 1.36 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr20_-_44496245 | 1.36 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr11_+_38280454 | 1.35 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr1_+_10118455 | 1.35 |
ENSDART00000152432
|
lratb.2
|
lecithin retinol acyltransferase b, tandem duplicate 2 |
| chr15_-_44601331 | 1.35 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr5_+_61301525 | 1.34 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr10_+_21786656 | 1.34 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr18_-_1185772 | 1.34 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr19_-_32710922 | 1.33 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr1_+_12766351 | 1.32 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr22_-_12160283 | 1.32 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr9_-_22821901 | 1.31 |
ENSDART00000101711
|
neb
|
nebulin |
| chr4_+_14900042 | 1.31 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr16_+_47207691 | 1.31 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr3_+_28953274 | 1.30 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr3_+_17537352 | 1.30 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr1_-_15797663 | 1.29 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr18_+_9637744 | 1.29 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
Network of associatons between targets according to the STRING database.
First level regulatory network of alx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.5 | 4.4 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.8 | 3.1 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.7 | 3.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.7 | 2.7 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.6 | 1.3 | GO:0022410 | circadian sleep/wake cycle process(GO:0022410) circadian sleep/wake cycle(GO:0042745) |
| 0.6 | 1.8 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.6 | 31.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.6 | 3.4 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.6 | 3.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.5 | 2.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.5 | 1.9 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.5 | 1.4 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.5 | 1.4 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.5 | 1.8 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.5 | 5.0 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.4 | 2.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.4 | 1.7 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.4 | 1.2 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.4 | 3.6 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.4 | 4.3 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.4 | 5.0 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.3 | 7.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 1.0 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.3 | 1.8 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.3 | 3.0 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.3 | 3.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.3 | 1.4 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.3 | 1.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.3 | 0.8 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.3 | 3.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 2.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.3 | 1.5 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.2 | 2.0 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 1.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 1.4 | GO:0070375 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.2 | 0.7 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.2 | 1.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 0.7 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.2 | 0.9 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 2.8 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 0.4 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.2 | 6.4 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.2 | 3.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 1.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 1.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.0 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.2 | 0.8 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 2.7 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.2 | 1.6 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.2 | 1.9 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 1.5 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.2 | 1.5 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 1.3 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.2 | 1.5 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.2 | 0.6 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.2 | 0.5 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.2 | 0.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 2.8 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 2.4 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.1 | 1.0 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.0 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.0 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 1.9 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.8 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 1.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 3.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 6.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.8 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 1.1 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 1.0 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 3.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 1.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.0 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.5 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.6 | GO:2000389 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 2.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.5 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 1.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 2.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 5.3 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 2.5 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 2.3 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.8 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 3.6 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 1.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 1.0 | GO:0046386 | deoxyribonucleotide catabolic process(GO:0009264) deoxyribose phosphate catabolic process(GO:0046386) |
| 0.1 | 1.6 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 5.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 2.8 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 3.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 1.5 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 0.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.4 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 1.8 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 2.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 1.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.2 | GO:1900158 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 3.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.0 | GO:0033198 | response to purine-containing compound(GO:0014074) response to ATP(GO:0033198) response to organophosphorus(GO:0046683) |
| 0.1 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 15.0 | GO:0050953 | sensory perception of light stimulus(GO:0050953) |
| 0.1 | 0.4 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 2.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 1.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 2.9 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 1.1 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.2 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 1.0 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.1 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 2.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.5 | GO:2000251 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 1.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 1.7 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.3 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 2.3 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 3.6 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 1.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.6 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0070296 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.8 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 2.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 2.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 1.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.6 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 1.2 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.3 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 1.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 2.1 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 1.2 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.5 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 2.3 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 3.2 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.3 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 1.7 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 1.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 1.4 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.8 | GO:0030641 | regulation of pH(GO:0006885) regulation of cellular pH(GO:0030641) |
| 0.0 | 0.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.1 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.3 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 2.2 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.0 | GO:0060347 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.0 | 2.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.0 | 0.3 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 0.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.7 | 31.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 3.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 5.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.4 | 4.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 2.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 2.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 2.6 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.3 | 1.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 3.0 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.3 | 1.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 5.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 9.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 3.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 1.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 0.5 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 3.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 5.0 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 6.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.8 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.9 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.1 | 1.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.5 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.9 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 10.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 3.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 27.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 5.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.5 | GO:0031968 | outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 11.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 1.3 | 5.1 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 1.0 | 3.1 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.7 | 2.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.6 | 3.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.5 | 2.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.5 | 2.7 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.4 | 4.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.4 | 1.3 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.4 | 3.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 2.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.4 | 1.2 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.4 | 5.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.4 | 4.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 5.0 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 1.8 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.4 | 2.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 2.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 3.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 3.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 2.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 2.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 2.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 2.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 2.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.3 | 1.4 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.3 | 1.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 2.6 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.3 | 1.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 0.8 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.3 | 2.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.9 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.4 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.2 | 2.0 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.2 | 2.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 3.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.8 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 1.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 1.8 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 0.8 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 5.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 2.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 3.0 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 0.8 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 5.0 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.1 | 1.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.9 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.5 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 2.2 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 1.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 5.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 6.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 4.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 3.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 3.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.0 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 3.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 4.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.7 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 2.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.0 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.5 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.0 | GO:0005231 | excitatory extracellular ligand-gated ion channel activity(GO:0005231) |
| 0.1 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 3.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.6 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.5 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.6 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 4.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 2.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.7 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.0 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.7 | GO:0005501 | retinoid binding(GO:0005501) isoprenoid binding(GO:0019840) |
| 0.0 | 1.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.9 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 2.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.9 | GO:0099589 | serotonin receptor activity(GO:0099589) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 2.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 1.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.9 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.2 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 1.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 3.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 1.9 | GO:0042626 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 6.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 3.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 2.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 6.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.0 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 1.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 5.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 3.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 3.1 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 2.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.5 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.0 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 2.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |