Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
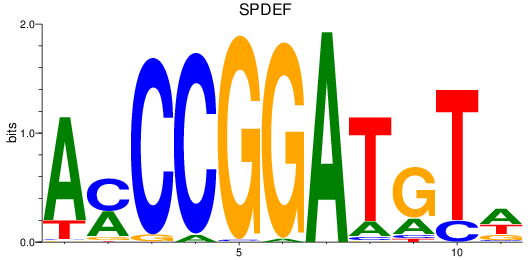
Results for SPDEF
Z-value: 1.70
Transcription factors associated with SPDEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPDEF
|
ENSDARG00000029930 | SAM pointed domain containing ETS transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPDEF | dr11_v1_chr6_-_54290227_54290227 | 0.63 | 4.2e-03 | Click! |
Activity profile of SPDEF motif
Sorted Z-values of SPDEF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_22749553 | 3.42 |
ENSDART00000040033
|
nup107
|
nucleoporin 107 |
| chr23_-_33775145 | 3.34 |
ENSDART00000132147
ENSDART00000027959 ENSDART00000160116 |
racgap1
|
Rac GTPase activating protein 1 |
| chr20_+_39344889 | 3.30 |
ENSDART00000009164
|
esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr20_+_13141408 | 3.22 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr8_+_23726708 | 3.17 |
ENSDART00000142395
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr18_+_29898955 | 3.08 |
ENSDART00000064080
|
cenpn
|
centromere protein N |
| chr6_-_34860574 | 3.02 |
ENSDART00000073957
|
slc35d1a
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1a |
| chr1_+_49435017 | 2.96 |
ENSDART00000124833
|
pdcd11
|
programmed cell death 11 |
| chr21_-_36619599 | 2.86 |
ENSDART00000065208
|
nop16
|
NOP16 nucleolar protein homolog (yeast) |
| chr18_-_3552414 | 2.70 |
ENSDART00000163762
ENSDART00000165434 ENSDART00000161197 ENSDART00000166841 ENSDART00000170260 |
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr19_+_34742706 | 2.64 |
ENSDART00000103276
|
fam206a
|
family with sequence similarity 206, member A |
| chr15_-_43625549 | 2.63 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr3_-_3398383 | 2.62 |
ENSDART00000047865
|
si:dkey-46g23.2
|
si:dkey-46g23.2 |
| chr17_-_12966907 | 2.60 |
ENSDART00000022874
|
psma6a
|
proteasome subunit alpha 6a |
| chr18_-_26797723 | 2.56 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr25_-_3347418 | 2.54 |
ENSDART00000082385
|
golt1bb
|
golgi transport 1Bb |
| chr3_-_34136778 | 2.52 |
ENSDART00000131951
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr14_-_23684814 | 2.51 |
ENSDART00000024604
|
mars2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr9_-_24218367 | 2.49 |
ENSDART00000135356
|
nabp1a
|
nucleic acid binding protein 1a |
| chr7_-_57637779 | 2.48 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr19_-_34742440 | 2.47 |
ENSDART00000122625
ENSDART00000175621 |
elp2
|
elongator acetyltransferase complex subunit 2 |
| chr9_+_56232548 | 2.46 |
ENSDART00000099276
|
cnot11
|
CCR4-NOT transcription complex, subunit 11 |
| chr11_+_2202987 | 2.45 |
ENSDART00000190008
ENSDART00000173139 |
hoxc6b
|
homeobox C6b |
| chr25_+_27405738 | 2.45 |
ENSDART00000183266
ENSDART00000115139 |
pot1
|
protection of telomeres 1 homolog |
| chr5_+_44806374 | 2.44 |
ENSDART00000184237
|
ctsla
|
cathepsin La |
| chr5_+_28497956 | 2.44 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr6_-_18531349 | 2.43 |
ENSDART00000160693
ENSDART00000169780 |
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr18_-_22735002 | 2.42 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr1_-_51720633 | 2.37 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr3_-_27065477 | 2.34 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr3_-_24681404 | 2.33 |
ENSDART00000161612
|
BX569789.1
|
|
| chr2_+_3044992 | 2.33 |
ENSDART00000020463
|
zgc:63882
|
zgc:63882 |
| chr19_+_9455218 | 2.32 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr16_+_33938227 | 2.29 |
ENSDART00000166254
|
gpn2
|
GPN-loop GTPase 2 |
| chr21_+_5080789 | 2.26 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr19_-_34089205 | 2.25 |
ENSDART00000163618
ENSDART00000161666 |
rp9
|
RP9, pre-mRNA splicing factor |
| chr8_+_28593707 | 2.24 |
ENSDART00000097213
|
tcf15
|
transcription factor 15 |
| chr11_-_16115804 | 2.24 |
ENSDART00000143436
ENSDART00000157928 |
rpf1
|
ribosome production factor 1 homolog |
| chr5_+_64856666 | 2.23 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr8_+_23726244 | 2.22 |
ENSDART00000132734
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr3_-_40276057 | 2.18 |
ENSDART00000132225
ENSDART00000074737 |
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr3_-_27066451 | 2.18 |
ENSDART00000156228
ENSDART00000156311 |
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr2_+_30182431 | 2.12 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr4_-_1824836 | 2.12 |
ENSDART00000111858
|
mrpl42
|
mitochondrial ribosomal protein L42 |
| chr12_+_6465557 | 2.11 |
ENSDART00000066477
ENSDART00000122271 |
dkk1b
|
dickkopf WNT signaling pathway inhibitor 1b |
| chr21_+_26539157 | 2.11 |
ENSDART00000021121
|
stx5al
|
syntaxin 5A, like |
| chr7_+_1505507 | 2.10 |
ENSDART00000161015
|
nop10
|
NOP10 ribonucleoprotein homolog (yeast) |
| chr1_+_24557414 | 2.10 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr7_-_30177691 | 2.09 |
ENSDART00000046689
|
tmed3
|
transmembrane p24 trafficking protein 3 |
| chr3_-_34136368 | 2.09 |
ENSDART00000136900
ENSDART00000186125 |
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr11_-_15296805 | 2.07 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr8_-_1267247 | 2.03 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr6_+_3334710 | 2.03 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr1_-_28950366 | 2.02 |
ENSDART00000110270
|
pwp2h
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr13_-_33700461 | 2.01 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr16_-_31824525 | 2.00 |
ENSDART00000058737
|
cdc42l
|
cell division cycle 42, like |
| chr5_-_29780752 | 2.00 |
ENSDART00000137400
ENSDART00000145021 |
cfap77
|
cilia and flagella associated protein 77 |
| chr3_+_19665319 | 1.99 |
ENSDART00000007857
ENSDART00000193509 |
mettl2a
|
methyltransferase like 2A |
| chr20_+_39685572 | 1.99 |
ENSDART00000050729
|
hddc2
|
HD domain containing 2 |
| chr12_-_31724198 | 1.97 |
ENSDART00000153056
ENSDART00000165299 ENSDART00000137464 ENSDART00000080173 |
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr21_-_40938382 | 1.96 |
ENSDART00000008593
|
yipf5
|
Yip1 domain family, member 5 |
| chr18_-_18584839 | 1.96 |
ENSDART00000159274
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr4_+_5506952 | 1.96 |
ENSDART00000032857
ENSDART00000160222 |
mapk11
|
mitogen-activated protein kinase 11 |
| chr14_-_10617923 | 1.95 |
ENSDART00000133723
ENSDART00000131939 ENSDART00000136649 |
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr24_+_19522094 | 1.95 |
ENSDART00000191042
ENSDART00000184714 ENSDART00000180782 |
sulf1
|
sulfatase 1 |
| chr22_-_10752471 | 1.93 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr15_-_15230264 | 1.92 |
ENSDART00000155400
|
rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr5_+_41477954 | 1.91 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr4_+_13931578 | 1.88 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr12_-_28363111 | 1.87 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr14_+_712115 | 1.86 |
ENSDART00000157494
ENSDART00000166516 ENSDART00000082017 ENSDART00000122374 ENSDART00000170203 |
ints10
|
integrator complex subunit 10 |
| chr2_+_48303142 | 1.85 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr8_+_47683539 | 1.84 |
ENSDART00000190701
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr13_+_33655404 | 1.83 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr9_-_28255029 | 1.82 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr14_+_4276394 | 1.82 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr16_-_44673851 | 1.82 |
ENSDART00000015139
|
dcaf13
|
ddb1 and cul4 associated factor 13 |
| chr5_+_51833132 | 1.80 |
ENSDART00000167491
|
papd4
|
PAP associated domain containing 4 |
| chr6_-_29159888 | 1.77 |
ENSDART00000110288
|
zbtb11
|
zinc finger and BTB domain containing 11 |
| chr1_+_12335816 | 1.77 |
ENSDART00000067086
|
nansa
|
N-acetylneuraminic acid synthase a |
| chr24_-_7826489 | 1.77 |
ENSDART00000112777
|
si:dkey-197c15.6
|
si:dkey-197c15.6 |
| chr22_+_10752787 | 1.75 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_16798923 | 1.74 |
ENSDART00000181603
ENSDART00000179816 ENSDART00000087107 ENSDART00000187009 |
eif4g1a
|
eukaryotic translation initiation factor 4 gamma, 1a |
| chr22_+_10752511 | 1.73 |
ENSDART00000081188
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_+_25084385 | 1.72 |
ENSDART00000134526
ENSDART00000111863 |
paxx
|
PAXX, non-homologous end joining factor |
| chr8_-_17184482 | 1.72 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr8_-_14080534 | 1.71 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr7_-_13906409 | 1.70 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr7_+_38809241 | 1.66 |
ENSDART00000190979
|
harbi1
|
harbinger transposase derived 1 |
| chr24_-_13349464 | 1.66 |
ENSDART00000134482
ENSDART00000139212 |
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr7_-_66693712 | 1.63 |
ENSDART00000021317
|
ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr5_-_52010122 | 1.63 |
ENSDART00000073627
ENSDART00000163898 ENSDART00000051003 |
cdk7
|
cyclin-dependent kinase 7 |
| chr22_-_5663354 | 1.62 |
ENSDART00000081774
|
ccdc51
|
coiled-coil domain containing 51 |
| chr5_+_44346691 | 1.61 |
ENSDART00000034523
|
tars
|
threonyl-tRNA synthetase |
| chr14_-_5407555 | 1.61 |
ENSDART00000001424
|
pcgf1
|
polycomb group ring finger 1 |
| chr22_+_5663529 | 1.59 |
ENSDART00000106141
|
tma7
|
translation machinery associated 7 homolog |
| chr1_-_31657644 | 1.59 |
ENSDART00000142296
|
dpcd
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr4_+_9536860 | 1.56 |
ENSDART00000130083
|
lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr9_-_41090048 | 1.54 |
ENSDART00000131681
ENSDART00000182552 |
asnsd1
|
asparagine synthetase domain containing 1 |
| chr20_-_48898560 | 1.54 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr6_-_18531760 | 1.54 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr13_+_9559461 | 1.54 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr23_-_24234371 | 1.53 |
ENSDART00000124539
|
ddost
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr5_+_42064144 | 1.53 |
ENSDART00000035235
|
si:ch211-202a12.4
|
si:ch211-202a12.4 |
| chr18_-_40884087 | 1.53 |
ENSDART00000059194
|
snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr8_-_41279326 | 1.53 |
ENSDART00000075491
|
pop5
|
POP5 homolog, ribonuclease P/MRP subunit |
| chr14_-_30897177 | 1.52 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr7_+_756942 | 1.51 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr14_-_43616572 | 1.49 |
ENSDART00000111189
|
gar1
|
GAR1 homolog, ribonucleoprotein |
| chr19_+_43004408 | 1.48 |
ENSDART00000038230
|
snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr1_+_41596099 | 1.47 |
ENSDART00000111367
|
si:dkey-56e3.3
|
si:dkey-56e3.3 |
| chr19_+_3842891 | 1.47 |
ENSDART00000159043
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr11_+_6374448 | 1.47 |
ENSDART00000135906
|
AL840638.2
|
|
| chr17_+_6452511 | 1.46 |
ENSDART00000064694
|
TATDN3
|
Danio rerio TatD DNase domain containing 3-like (LOC571912), mRNA. |
| chr23_+_16928506 | 1.46 |
ENSDART00000162292
ENSDART00000080625 |
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr16_+_33931032 | 1.46 |
ENSDART00000167240
|
snip1
|
Smad nuclear interacting protein |
| chr17_-_31579715 | 1.44 |
ENSDART00000110167
ENSDART00000191092 |
rpap1
|
RNA polymerase II associated protein 1 |
| chr7_-_71384391 | 1.43 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr5_+_47863153 | 1.43 |
ENSDART00000051518
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr2_+_32846602 | 1.41 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr4_-_20108833 | 1.40 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr21_-_30994577 | 1.40 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr18_+_6638726 | 1.40 |
ENSDART00000142755
ENSDART00000167781 |
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr15_-_43284021 | 1.39 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr12_+_46543572 | 1.39 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr13_-_33654931 | 1.37 |
ENSDART00000020350
|
snx5
|
sorting nexin 5 |
| chr15_+_17343319 | 1.37 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr5_-_28041715 | 1.36 |
ENSDART00000078660
|
zgc:113436
|
zgc:113436 |
| chr5_-_25084318 | 1.36 |
ENSDART00000089339
|
dph7
|
diphthamide biosynthesis 7 |
| chr3_+_59880317 | 1.34 |
ENSDART00000166922
ENSDART00000108647 |
alyref
|
Aly/REF export factor |
| chr6_-_54444929 | 1.32 |
ENSDART00000154121
|
sys1
|
Sys1 golgi trafficking protein |
| chr23_+_4260458 | 1.31 |
ENSDART00000103747
|
srsf6a
|
serine/arginine-rich splicing factor 6a |
| chr1_-_44581937 | 1.30 |
ENSDART00000009858
|
tmx2b
|
thioredoxin-related transmembrane protein 2b |
| chr9_+_25853052 | 1.30 |
ENSDART00000127135
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr15_-_34668485 | 1.30 |
ENSDART00000186605
|
bag6
|
BCL2 associated athanogene 6 |
| chr2_-_45510699 | 1.29 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr6_-_2222707 | 1.28 |
ENSDART00000022179
|
prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr20_-_5369105 | 1.28 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr12_-_14211293 | 1.28 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr17_+_24684778 | 1.27 |
ENSDART00000146309
ENSDART00000082237 |
znf593
|
zinc finger protein 593 |
| chr10_+_15970 | 1.27 |
ENSDART00000040240
|
tiprl
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr24_-_13349802 | 1.26 |
ENSDART00000164729
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr18_+_6638974 | 1.25 |
ENSDART00000162398
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr22_-_16494406 | 1.25 |
ENSDART00000062727
|
stx6
|
syntaxin 6 |
| chr7_-_34062301 | 1.25 |
ENSDART00000052404
|
map2k5
|
mitogen-activated protein kinase kinase 5 |
| chr6_-_54433995 | 1.24 |
ENSDART00000017230
|
snrpc
|
small nuclear ribonucleoprotein polypeptide C |
| chr5_+_29714786 | 1.24 |
ENSDART00000148314
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr6_+_3334392 | 1.23 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr3_-_36419641 | 1.23 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr2_-_3044947 | 1.21 |
ENSDART00000192642
|
guk1a
|
guanylate kinase 1a |
| chr25_+_3347461 | 1.21 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr6_+_41808673 | 1.20 |
ENSDART00000038163
|
rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr9_+_2333927 | 1.18 |
ENSDART00000123340
|
atp5mc3a
|
ATP synthase membrane subunit c locus 3a |
| chr5_+_32490238 | 1.17 |
ENSDART00000191839
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr21_-_22892124 | 1.17 |
ENSDART00000065563
|
ccdc90b
|
coiled-coil domain containing 90B |
| chr9_+_40874194 | 1.17 |
ENSDART00000141548
|
hibch
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr16_+_38337783 | 1.15 |
ENSDART00000135008
|
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
| chr14_+_28438947 | 1.14 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr5_+_51833305 | 1.13 |
ENSDART00000165276
ENSDART00000166443 |
papd4
|
PAP associated domain containing 4 |
| chr17_+_8754020 | 1.13 |
ENSDART00000105322
|
edrf1
|
erythroid differentiation regulatory factor 1 |
| chr7_-_29356084 | 1.13 |
ENSDART00000075757
|
gtf2a2
|
general transcription factor IIA, 2 |
| chr1_+_46598764 | 1.13 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr15_+_7064819 | 1.12 |
ENSDART00000155268
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr3_-_30153242 | 1.12 |
ENSDART00000077089
|
nucb1
|
nucleobindin 1 |
| chr11_+_24348425 | 1.12 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr1_-_23294753 | 1.10 |
ENSDART00000013263
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr23_-_4225606 | 1.10 |
ENSDART00000014152
ENSDART00000133341 |
aar2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr25_+_8921425 | 1.10 |
ENSDART00000128591
|
accs
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| chr2_+_2772447 | 1.09 |
ENSDART00000124882
|
thoc1
|
THO complex 1 |
| chr5_-_67292690 | 1.07 |
ENSDART00000062366
|
rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr20_-_16906623 | 1.06 |
ENSDART00000012859
ENSDART00000171628 |
psma6b
|
proteasome subunit alpha 6b |
| chr11_+_44622472 | 1.06 |
ENSDART00000159068
ENSDART00000166323 ENSDART00000187753 |
rbm34
|
RNA binding motif protein 34 |
| chr17_+_20237727 | 1.06 |
ENSDART00000180115
|
smndc1
|
survival motor neuron domain containing 1 |
| chr5_+_64397454 | 1.06 |
ENSDART00000015940
|
edf1
|
endothelial differentiation-related factor 1 |
| chr13_-_5569562 | 1.05 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr8_-_1266181 | 1.05 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr9_-_41090343 | 1.04 |
ENSDART00000187769
ENSDART00000180078 ENSDART00000166785 |
asnsd1
|
asparagine synthetase domain containing 1 |
| chr5_+_54555567 | 1.04 |
ENSDART00000171159
|
anapc2
|
anaphase promoting complex subunit 2 |
| chr20_-_23254876 | 1.03 |
ENSDART00000141510
|
ociad1
|
OCIA domain containing 1 |
| chr5_-_26893310 | 1.02 |
ENSDART00000126669
|
lman2lb
|
lectin, mannose-binding 2-like b |
| chr20_-_10487951 | 1.02 |
ENSDART00000064112
|
glrx5
|
glutaredoxin 5 homolog (S. cerevisiae) |
| chr23_+_36115541 | 1.02 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr15_+_38299385 | 1.01 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr20_+_27087539 | 1.01 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr8_+_24740013 | 1.01 |
ENSDART00000126897
|
lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr19_+_37118547 | 0.97 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr7_+_35193832 | 0.95 |
ENSDART00000189002
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr9_-_40873934 | 0.95 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr7_+_29177191 | 0.94 |
ENSDART00000008096
|
aph1b
|
APH1B gamma secretase subunit |
| chr22_-_10826 | 0.94 |
ENSDART00000125700
|
mrpl20
|
mitochondrial ribosomal protein L20 |
| chr15_-_29114449 | 0.94 |
ENSDART00000145748
ENSDART00000109482 ENSDART00000179123 |
zgc:162698
|
zgc:162698 |
| chr2_-_42173834 | 0.94 |
ENSDART00000098357
ENSDART00000144707 |
slc39a6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr15_-_2519640 | 0.93 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr4_+_13931733 | 0.93 |
ENSDART00000141742
ENSDART00000067175 |
pphln1
|
periphilin 1 |
| chr3_-_30152836 | 0.92 |
ENSDART00000165920
|
nucb1
|
nucleobindin 1 |
| chr5_+_29715040 | 0.92 |
ENSDART00000192563
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr20_+_52492192 | 0.91 |
ENSDART00000057986
|
TSTA3 (1 of many)
|
zgc:100864 |
| chr17_-_24684687 | 0.91 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr7_-_38861741 | 0.90 |
ENSDART00000173629
ENSDART00000037361 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr1_-_44048798 | 0.90 |
ENSDART00000073746
|
si:ch73-109d9.2
|
si:ch73-109d9.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPDEF
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 1.1 | 3.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.8 | 2.4 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.7 | 2.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.7 | 2.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.7 | 2.1 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.6 | 2.6 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.6 | 1.8 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.6 | 1.8 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.6 | 2.8 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.5 | 2.7 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.5 | 2.5 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.5 | 1.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.4 | 2.1 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.4 | 3.2 | GO:0072422 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.4 | 2.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 1.8 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.3 | 5.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.3 | 1.9 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.3 | 1.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 2.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 0.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 1.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 8.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.3 | 1.5 | GO:0090467 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.2 | 2.0 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 1.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 2.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 3.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 1.5 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.2 | 1.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.2 | 0.8 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.2 | 3.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 1.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 1.1 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.2 | 0.6 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.2 | 0.7 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 2.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 4.6 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.2 | 1.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 2.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 0.7 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.2 | 0.9 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 1.1 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.2 | 3.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 3.1 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.4 | GO:0015808 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.1 | 0.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.6 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 2.5 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 3.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.8 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 1.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.9 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 2.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 2.5 | GO:0031577 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.1 | 1.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.3 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 3.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.8 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 1.6 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.1 | 1.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 2.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.5 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 2.8 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 2.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.6 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.1 | 1.7 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.8 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 2.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 1.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 2.2 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 1.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 2.0 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.6 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 3.3 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 1.4 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 1.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.2 | GO:0006513 | postreplication repair(GO:0006301) protein monoubiquitination(GO:0006513) |
| 0.1 | 0.9 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 4.6 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 2.8 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 2.5 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.2 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.0 | 1.0 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 1.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.9 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.4 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.4 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.5 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 3.6 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 1.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 2.2 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 3.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.1 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 2.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 1.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 1.2 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 2.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.7 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 1.6 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 1.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 1.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 2.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.8 | GO:0016051 | carbohydrate biosynthetic process(GO:0016051) |
| 0.0 | 0.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.3 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 4.7 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.9 | 3.6 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.9 | 6.0 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.8 | 2.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.6 | 1.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.6 | 2.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.6 | 2.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.5 | 5.0 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.4 | 2.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 1.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.4 | 1.5 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.3 | 1.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 3.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.3 | 2.3 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.3 | 3.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 1.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 1.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 3.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 2.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 2.0 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.2 | 1.7 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 1.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 0.8 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 1.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 4.1 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.2 | 2.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 1.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 1.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 2.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.2 | 1.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 0.6 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.2 | 4.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 1.6 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 0.7 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 3.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.5 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 2.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 2.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 1.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.9 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 2.3 | GO:0030684 | preribosome(GO:0030684) |
| 0.1 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 3.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 3.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 3.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 2.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 2.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 2.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 4.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.5 | GO:0000785 | chromatin(GO:0000785) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 1.0 | 2.9 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.7 | 2.2 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.6 | 2.6 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.6 | 2.5 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.6 | 1.8 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.6 | 2.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.6 | 2.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.5 | 2.6 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.5 | 4.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.5 | 2.0 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.5 | 3.3 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.4 | 2.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 1.5 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.4 | 2.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 2.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 5.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.3 | 1.5 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.3 | 1.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.3 | 1.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.2 | 1.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 2.4 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 1.8 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.2 | 1.5 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 2.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 0.8 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 1.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 1.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 0.5 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.2 | 2.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.9 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.6 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 2.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.9 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.9 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 3.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.5 | GO:0070035 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 2.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 2.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 2.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.5 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.4 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 1.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 1.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.0 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 3.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.2 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.1 | 0.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 2.3 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.1 | 0.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 2.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.2 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 3.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 3.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 4.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.8 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 2.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 1.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 3.3 | GO:0016407 | acetyltransferase activity(GO:0016407) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 2.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.0 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 1.7 | GO:0004518 | nuclease activity(GO:0004518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 2.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 5.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 6.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.4 | 5.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.4 | 1.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 2.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 1.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 2.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 3.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 1.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 3.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.5 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 5.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.4 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.1 | 2.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 0.9 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 3.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 2.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.6 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.1 | 1.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 3.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 0.7 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.1 | 1.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.4 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.2 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |